+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: PDB / ID: 9hfl | ||||||
|---|---|---|---|---|---|---|---|
| タイトル | Cryo-EM structure of the human snRNA export complex comprising CBC-PHAX-CRM1-RanGTP and capped-RNA | ||||||
 要素 要素 |
| ||||||
 キーワード キーワード | RNA BINDING PROTEIN / snRNA export / Exportin / Cap-binding / Co-transcriptional regulation | ||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報mRNA cap binding complex binding / positive regulation of RNA binding / snRNA export from nucleus / nuclear cap binding complex / histone mRNA metabolic process / RNA cap binding complex / mRNA metabolic process / cellular response to triglyceride / RNA stabilization / cellular response to salt ...mRNA cap binding complex binding / positive regulation of RNA binding / snRNA export from nucleus / nuclear cap binding complex / histone mRNA metabolic process / RNA cap binding complex / mRNA metabolic process / cellular response to triglyceride / RNA stabilization / cellular response to salt / positive regulation of RNA export from nucleus / HuR (ELAVL1) binds and stabilizes mRNA / positive regulation of mRNA 3'-end processing / cap-dependent translational initiation / annulate lamellae / Processing of Intronless Pre-mRNAs / regulation of proteasomal ubiquitin-dependent protein catabolic process / RNA cap binding / snRNA binding / pre-miRNA export from nucleus / RNA nuclear export complex / snRNA import into nucleus / regulation of mRNA processing / primary miRNA processing / miRNA-mediated post-transcriptional gene silencing / manchette / nuclear export signal receptor activity / regulation of centrosome duplication / cellular response to mineralocorticoid stimulus / SLBP independent Processing of Histone Pre-mRNAs / SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs / regulatory ncRNA-mediated post-transcriptional gene silencing / Regulation of cholesterol biosynthesis by SREBP (SREBF) / RNA 7-methylguanosine cap binding / Transport of the SLBP independent Mature mRNA / importin-alpha family protein binding / Transport of the SLBP Dependant Mature mRNA / alternative mRNA splicing, via spliceosome / regulation of protein export from nucleus / mRNA 3'-end processing / Transport of Mature mRNA Derived from an Intronless Transcript / positive regulation of mRNA splicing, via spliceosome / Rev-mediated nuclear export of HIV RNA / Nuclear import of Rev protein / mRNA 3'-end processing / protein localization to nucleolus / mRNA cis splicing, via spliceosome / NEP/NS2 Interacts with the Cellular Export Machinery / Transport of Mature mRNA derived from an Intron-Containing Transcript / tRNA processing in the nucleus / GTP metabolic process / RNA catabolic process / Postmitotic nuclear pore complex (NPC) reformation / RNA Polymerase II Transcription Termination / Abortive elongation of HIV-1 transcript in the absence of Tat / nuclear-transcribed mRNA catabolic process, nonsense-mediated decay / regulation of translational initiation / FGFR2 alternative splicing / nucleocytoplasmic transport / MicroRNA (miRNA) biogenesis / Signaling by FGFR2 IIIa TM / DNA metabolic process / Formation of the Early Elongation Complex / Formation of the HIV-1 Early Elongation Complex / mRNA Capping / Maturation of hRSV A proteins / spliceosomal complex assembly / mRNA Splicing - Minor Pathway / dynein intermediate chain binding / Processing of Capped Intron-Containing Pre-mRNA / mitotic sister chromatid segregation / RNA polymerase II transcribes snRNA genes / 7-methylguanosine mRNA capping / viral process / spermatid development / ribosomal large subunit export from nucleus / Estrogen-dependent nuclear events downstream of ESR-membrane signaling / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / protein localization to nucleus / positive regulation of protein binding / sperm flagellum / toxic substance binding / nuclear pore / mRNA export from nucleus / Formation of HIV-1 elongation complex containing HIV-1 Tat / Cajal body / Formation of HIV elongation complex in the absence of HIV Tat / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / ribosomal subunit export from nucleus / Cyclin A/B1/B2 associated events during G2/M transition / Formation of RNA Pol II elongation complex / ribosomal small subunit export from nucleus / NPAS4 regulates expression of target genes / Amplification of signal from unattached kinetochores via a MAD2 inhibitory signal / RNA Polymerase II Pre-transcription Events / centriole / Transcriptional and post-translational regulation of MITF-M expression and activity / Mitotic Prometaphase / mRNA Splicing - Major Pathway / EML4 and NUDC in mitotic spindle formation 類似検索 - 分子機能 | ||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) | ||||||
| 手法 | 電子顕微鏡法 / 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 2.62 Å | ||||||
 データ登録者 データ登録者 | Dubiez, E. / Cusack, S. / Kadlec, J. | ||||||
| 資金援助 |  フランス, 1件 フランス, 1件
| ||||||
 引用 引用 |  ジャーナル: Nat Struct Mol Biol / 年: 2025 ジャーナル: Nat Struct Mol Biol / 年: 2025タイトル: Structural basis for the synergistic assembly of the snRNA export complex. 著者: Etienne Dubiez / William Garland / Maja Finderup Brask / Elisabetta Boeri Erba / Torben Heick Jensen / Jan Kadlec / Stephen Cusack /   要旨: The nuclear cap-binding complex (CBC) and its partner Arsenite-Resistance Protein 2 (ARS2) regulate the fate of RNA polymerase II transcripts via mutually exclusive interactions with RNA effectors. ...The nuclear cap-binding complex (CBC) and its partner Arsenite-Resistance Protein 2 (ARS2) regulate the fate of RNA polymerase II transcripts via mutually exclusive interactions with RNA effectors. One such effector is PHAX, which mediates the nuclear export of U-rich small nuclear RNAs (snRNAs). Here we present the cryo-electron microscopy structure of the human snRNA export complex comprising phosphorylated PHAX, CBC, CRM1-RanGTP and capped RNA. The central region of PHAX bridges CBC to the export factor CRM1-RanGTP, while also reinforcing cap dinucleotide binding. Additionally, PHAX interacts with a distant region of CRM1, facilitating contacts of the essential phosphorylated region of PHAX with the prominent basic surface of RanGTP. CBC engagement within the snRNA export complex is incompatible with its binding to other RNA effectors such as ALYREF or NCBP3. We demonstrate that snRNA export complex formation requires synergistic binding of all its components, which in turn displaces ARS2 from CBC and commits the complex for export. | ||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 構造ビューア | 分子:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- ダウンロードとリンク
ダウンロードとリンク
- ダウンロード
ダウンロード
| PDBx/mmCIF形式 |  9hfl.cif.gz 9hfl.cif.gz | 860.2 KB | 表示 |  PDBx/mmCIF形式 PDBx/mmCIF形式 |
|---|---|---|---|---|
| PDB形式 |  pdb9hfl.ent.gz pdb9hfl.ent.gz | 700.7 KB | 表示 |  PDB形式 PDB形式 |
| PDBx/mmJSON形式 |  9hfl.json.gz 9hfl.json.gz | ツリー表示 |  PDBx/mmJSON形式 PDBx/mmJSON形式 | |
| その他 |  その他のダウンロード その他のダウンロード |
-検証レポート
| 文書・要旨 |  9hfl_validation.pdf.gz 9hfl_validation.pdf.gz | 1.5 MB | 表示 |  wwPDB検証レポート wwPDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  9hfl_full_validation.pdf.gz 9hfl_full_validation.pdf.gz | 1.5 MB | 表示 | |
| XML形式データ |  9hfl_validation.xml.gz 9hfl_validation.xml.gz | 69.7 KB | 表示 | |
| CIF形式データ |  9hfl_validation.cif.gz 9hfl_validation.cif.gz | 107 KB | 表示 | |
| アーカイブディレクトリ |  https://data.pdbj.org/pub/pdb/validation_reports/hf/9hfl https://data.pdbj.org/pub/pdb/validation_reports/hf/9hfl ftp://data.pdbj.org/pub/pdb/validation_reports/hf/9hfl ftp://data.pdbj.org/pub/pdb/validation_reports/hf/9hfl | HTTPS FTP |
-関連構造データ
| 関連構造データ |  52115MC M: このデータのモデリングに利用したマップデータ C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
- 集合体
集合体
| 登録構造単位 | 
|
|---|---|
| 1 |
|
- 要素
要素
-タンパク質 , 3種, 4分子 ABNP
| #1: タンパク質 | 分子量: 123518.359 Da / 分子数: 1 / 由来タイプ: 組換発現 / 詳細: Human Exportin 1 CRM1 / 由来: (組換発現)  Homo sapiens (ヒト) / 遺伝子: XPO1, CRM1 / 発現宿主: Homo sapiens (ヒト) / 遺伝子: XPO1, CRM1 / 発現宿主:  |
|---|---|
| #2: タンパク質 | 分子量: 24441.135 Da / 分子数: 1 / Mutation: Q69L / 由来タイプ: 組換発現 / 詳細: RanGTP Q69L catalytic mutant / 由来: (組換発現)  Homo sapiens (ヒト) / 遺伝子: RAN, ARA24, OK/SW-cl.81 / 発現宿主: Homo sapiens (ヒト) / 遺伝子: RAN, ARA24, OK/SW-cl.81 / 発現宿主:  参照: UniProt: P62826, 加水分解酵素; 酸無水物に作用; GTPに作用・細胞または細胞小器官の運動に関与 |
| #5: タンパク質 | 分子量: 44468.574 Da / 分子数: 2 / 由来タイプ: 組換発現 / 由来: (組換発現)  Homo sapiens (ヒト) / 遺伝子: PHAX, RNUXA / 発現宿主: Homo sapiens (ヒト) / 遺伝子: PHAX, RNUXA / 発現宿主:  |
-Nuclear cap-binding protein subunit ... , 2種, 2分子 CD
| #3: タンパク質 | 分子量: 91960.297 Da / 分子数: 1 / 由来タイプ: 組換発現 / 詳細: Human NCBP1 CBP80 / 由来: (組換発現)  Homo sapiens (ヒト) / 遺伝子: NCBP1, CBP80, NCBP Homo sapiens (ヒト) / 遺伝子: NCBP1, CBP80, NCBP発現宿主:  Baculovirus expression vector pFastBac1-HM (ウイルス) Baculovirus expression vector pFastBac1-HM (ウイルス)参照: UniProt: Q09161 |
|---|---|
| #4: タンパク質 | 分子量: 18028.131 Da / 分子数: 1 / 由来タイプ: 組換発現 / 詳細: Human NCBP2 CBP20 / 由来: (組換発現)  Homo sapiens (ヒト) / 遺伝子: NCBP2, CBP20, PIG55 / 発現宿主: Homo sapiens (ヒト) / 遺伝子: NCBP2, CBP20, PIG55 / 発現宿主:  |
-RNA鎖 , 1種, 1分子 R
| #6: RNA鎖 | 分子量: 4439.715 Da / 分子数: 1 / 由来タイプ: 合成 / 由来: (合成)  Homo sapiens (ヒト) Homo sapiens (ヒト) |
|---|
-非ポリマー , 4種, 35分子 

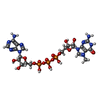




| #7: 化合物 | ChemComp-GTP / |
|---|---|
| #8: 化合物 | ChemComp-MG / |
| #9: 化合物 | ChemComp-GTA / |
| #10: 水 | ChemComp-HOH / |
-詳細
| 研究の焦点であるリガンドがあるか | Y |
|---|---|
| Has protein modification | N |
-実験情報
-実験
| 実験 | 手法: 電子顕微鏡法 |
|---|---|
| EM実験 | 試料の集合状態: PARTICLE / 3次元再構成法: 単粒子再構成法 |
- 試料調製
試料調製
| 構成要素 | 名称: Human snRNA export complex comprising CBC-PHAX-CRM1-RanGTP and capped-RNA タイプ: COMPLEX / Entity ID: #1-#6 / 由来: RECOMBINANT |
|---|---|
| 分子量 | 実験値: NO |
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 由来(組換発現) | 生物種:  |
| 緩衝液 | pH: 8 |
| 試料 | 濃度: 1 mg/ml / 包埋: NO / シャドウイング: NO / 染色: NO / 凍結: YES |
| 急速凍結 | 凍結剤: ETHANE |
- 電子顕微鏡撮影
電子顕微鏡撮影
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
|---|---|
| 顕微鏡 | モデル: TFS KRIOS |
| 電子銃 | 電子線源:  FIELD EMISSION GUN / 加速電圧: 300 kV / 照射モード: FLOOD BEAM FIELD EMISSION GUN / 加速電圧: 300 kV / 照射モード: FLOOD BEAM |
| 電子レンズ | モード: BRIGHT FIELD / 最大 デフォーカス(公称値): 2200 nm / 最小 デフォーカス(公称値): 1000 nm |
| 撮影 | 電子線照射量: 40.5 e/Å2 / フィルム・検出器のモデル: GATAN K3 (6k x 4k) |
- 解析
解析
| EMソフトウェア | 名称: PHENIX / カテゴリ: モデル精密化 | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF補正 | タイプ: NONE | ||||||||||||||||||||||||
| 3次元再構成 | 解像度: 2.62 Å / 解像度の算出法: FSC 0.143 CUT-OFF / 粒子像の数: 202738 / 対称性のタイプ: POINT | ||||||||||||||||||||||||
| 拘束条件 |
|
 ムービー
ムービー コントローラー
コントローラー



 PDBj
PDBj












































