[English] 日本語
 Yorodumi
Yorodumi- PDB-9hfl: Cryo-EM structure of the human snRNA export complex comprising CB... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 9hfl | ||||||
|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the human snRNA export complex comprising CBC-PHAX-CRM1-RanGTP and capped-RNA | ||||||
 Components Components |
| ||||||
 Keywords Keywords | RNA BINDING PROTEIN / snRNA export / Exportin / Cap-binding / Co-transcriptional regulation | ||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of RNA binding / mRNA cap binding complex binding / snRNA export from nucleus / nuclear cap binding complex / histone mRNA metabolic process / RNA cap binding complex / mRNA metabolic process / cellular response to triglyceride / RNA stabilization / cellular response to salt ...positive regulation of RNA binding / mRNA cap binding complex binding / snRNA export from nucleus / nuclear cap binding complex / histone mRNA metabolic process / RNA cap binding complex / mRNA metabolic process / cellular response to triglyceride / RNA stabilization / cellular response to salt / positive regulation of RNA export from nucleus / HuR (ELAVL1) binds and stabilizes mRNA / positive regulation of mRNA 3'-end processing / cap-dependent translational initiation / annulate lamellae / Processing of Intronless Pre-mRNAs / regulation of proteasomal ubiquitin-dependent protein catabolic process / RNA cap binding / snRNA binding / pre-miRNA export from nucleus / RNA nuclear export complex / snRNA import into nucleus / regulation of mRNA processing / primary miRNA processing / miRNA-mediated post-transcriptional gene silencing / manchette / nuclear export signal receptor activity / regulation of centrosome duplication / cellular response to mineralocorticoid stimulus / SLBP independent Processing of Histone Pre-mRNAs / SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs / regulatory ncRNA-mediated post-transcriptional gene silencing / Regulation of cholesterol biosynthesis by SREBP (SREBF) / Transport of the SLBP independent Mature mRNA / RNA 7-methylguanosine cap binding / Transport of the SLBP Dependant Mature mRNA / alternative mRNA splicing, via spliceosome / importin-alpha family protein binding / positive regulation of mRNA splicing, via spliceosome / mRNA 3'-end processing / Transport of Mature mRNA Derived from an Intronless Transcript / regulation of protein export from nucleus / Rev-mediated nuclear export of HIV RNA / Nuclear import of Rev protein / mRNA 3'-end processing / mRNA cis splicing, via spliceosome / protein localization to nucleolus / NEP/NS2 Interacts with the Cellular Export Machinery / Transport of Mature mRNA derived from an Intron-Containing Transcript / tRNA processing in the nucleus / GTP metabolic process / Postmitotic nuclear pore complex (NPC) reformation / RNA catabolic process / Abortive elongation of HIV-1 transcript in the absence of Tat / RNA Polymerase II Transcription Termination / nuclear-transcribed mRNA catabolic process, nonsense-mediated decay / regulation of translational initiation / FGFR2 alternative splicing / nucleocytoplasmic transport / MicroRNA (miRNA) biogenesis / Signaling by FGFR2 IIIa TM / DNA metabolic process / Formation of the Early Elongation Complex / Formation of the HIV-1 Early Elongation Complex / mRNA Capping / spliceosomal complex assembly / mRNA Splicing - Minor Pathway / dynein intermediate chain binding / Maturation of hRSV A proteins / Processing of Capped Intron-Containing Pre-mRNA / mitotic sister chromatid segregation / viral process / RNA polymerase II transcribes snRNA genes / 7-methylguanosine mRNA capping / spermatid development / ribosomal large subunit export from nucleus / Estrogen-dependent nuclear events downstream of ESR-membrane signaling / protein localization to nucleus / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / toxic substance binding / nuclear pore / sperm flagellum / positive regulation of protein binding / mRNA export from nucleus / Formation of HIV-1 elongation complex containing HIV-1 Tat / Cajal body / Formation of HIV elongation complex in the absence of HIV Tat / Cyclin A/B1/B2 associated events during G2/M transition / ribosomal subunit export from nucleus / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / Formation of RNA Pol II elongation complex / NPAS4 regulates expression of target genes / ribosomal small subunit export from nucleus / Amplification of signal from unattached kinetochores via a MAD2 inhibitory signal / RNA Polymerase II Pre-transcription Events / centriole / Transcriptional and post-translational regulation of MITF-M expression and activity / mRNA Splicing - Major Pathway / Mitotic Prometaphase / EML4 and NUDC in mitotic spindle formation Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 2.62 Å | ||||||
 Authors Authors | Dubiez, E. / Cusack, S. / Kadlec, J. | ||||||
| Funding support |  France, 1items France, 1items
| ||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2025 Journal: Nat Struct Mol Biol / Year: 2025Title: Structural basis for the synergistic assembly of the snRNA export complex. Authors: Etienne Dubiez / William Garland / Maja Finderup Brask / Elisabetta Boeri Erba / Torben Heick Jensen / Jan Kadlec / Stephen Cusack /   Abstract: The nuclear cap-binding complex (CBC) and its partner Arsenite-Resistance Protein 2 (ARS2) regulate the fate of RNA polymerase II transcripts via mutually exclusive interactions with RNA effectors. ...The nuclear cap-binding complex (CBC) and its partner Arsenite-Resistance Protein 2 (ARS2) regulate the fate of RNA polymerase II transcripts via mutually exclusive interactions with RNA effectors. One such effector is PHAX, which mediates the nuclear export of U-rich small nuclear RNAs (snRNAs). Here we present the cryo-electron microscopy structure of the human snRNA export complex comprising phosphorylated PHAX, CBC, CRM1-RanGTP and capped RNA. The central region of PHAX bridges CBC to the export factor CRM1-RanGTP, while also reinforcing cap dinucleotide binding. Additionally, PHAX interacts with a distant region of CRM1, facilitating contacts of the essential phosphorylated region of PHAX with the prominent basic surface of RanGTP. CBC engagement within the snRNA export complex is incompatible with its binding to other RNA effectors such as ALYREF or NCBP3. We demonstrate that snRNA export complex formation requires synergistic binding of all its components, which in turn displaces ARS2 from CBC and commits the complex for export. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  9hfl.cif.gz 9hfl.cif.gz | 860.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb9hfl.ent.gz pdb9hfl.ent.gz | 700.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  9hfl.json.gz 9hfl.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  9hfl_validation.pdf.gz 9hfl_validation.pdf.gz | 1.5 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  9hfl_full_validation.pdf.gz 9hfl_full_validation.pdf.gz | 1.5 MB | Display | |
| Data in XML |  9hfl_validation.xml.gz 9hfl_validation.xml.gz | 69.7 KB | Display | |
| Data in CIF |  9hfl_validation.cif.gz 9hfl_validation.cif.gz | 107 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/hf/9hfl https://data.pdbj.org/pub/pdb/validation_reports/hf/9hfl ftp://data.pdbj.org/pub/pdb/validation_reports/hf/9hfl ftp://data.pdbj.org/pub/pdb/validation_reports/hf/9hfl | HTTPS FTP |
-Related structure data
| Related structure data |  52115MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Protein , 3 types, 4 molecules ABNP
| #1: Protein | Mass: 123518.359 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Details: Human Exportin 1 CRM1 / Source: (gene. exp.)  Homo sapiens (human) / Gene: XPO1, CRM1 / Production host: Homo sapiens (human) / Gene: XPO1, CRM1 / Production host:  |
|---|---|
| #2: Protein | Mass: 24441.135 Da / Num. of mol.: 1 / Mutation: Q69L Source method: isolated from a genetically manipulated source Details: RanGTP Q69L catalytic mutant / Source: (gene. exp.)  Homo sapiens (human) / Gene: RAN, ARA24, OK/SW-cl.81 / Production host: Homo sapiens (human) / Gene: RAN, ARA24, OK/SW-cl.81 / Production host:  References: UniProt: P62826, Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement |
| #5: Protein | Mass: 44468.574 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: PHAX, RNUXA / Production host: Homo sapiens (human) / Gene: PHAX, RNUXA / Production host:  |
-Nuclear cap-binding protein subunit ... , 2 types, 2 molecules CD
| #3: Protein | Mass: 91960.297 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Details: Human NCBP1 CBP80 / Source: (gene. exp.)  Homo sapiens (human) / Gene: NCBP1, CBP80, NCBP / Production host: Homo sapiens (human) / Gene: NCBP1, CBP80, NCBP / Production host:  Baculovirus expression vector pFastBac1-HM / References: UniProt: Q09161 Baculovirus expression vector pFastBac1-HM / References: UniProt: Q09161 |
|---|---|
| #4: Protein | Mass: 18028.131 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Details: Human NCBP2 CBP20 / Source: (gene. exp.)  Homo sapiens (human) / Gene: NCBP2, CBP20, PIG55 / Production host: Homo sapiens (human) / Gene: NCBP2, CBP20, PIG55 / Production host:  |
-RNA chain , 1 types, 1 molecules R
| #6: RNA chain | Mass: 4439.715 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.)  Homo sapiens (human) Homo sapiens (human) |
|---|
-Non-polymers , 4 types, 35 molecules 

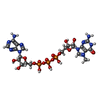




| #7: Chemical | ChemComp-GTP / |
|---|---|
| #8: Chemical | ChemComp-MG / |
| #9: Chemical | ChemComp-GTA / |
| #10: Water | ChemComp-HOH / |
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | N |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Human snRNA export complex comprising CBC-PHAX-CRM1-RanGTP and capped-RNA Type: COMPLEX / Entity ID: #1-#6 / Source: RECOMBINANT |
|---|---|
| Molecular weight | Experimental value: NO |
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Source (recombinant) | Organism:  |
| Buffer solution | pH: 8 |
| Specimen | Conc.: 1 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: TFS KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal defocus max: 2200 nm / Nominal defocus min: 1000 nm |
| Image recording | Electron dose: 40.5 e/Å2 / Film or detector model: GATAN K3 (6k x 4k) |
- Processing
Processing
| EM software | Name: PHENIX / Category: model refinement | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: NONE | ||||||||||||||||||||||||
| 3D reconstruction | Resolution: 2.62 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 202738 / Symmetry type: POINT | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller


 PDBj
PDBj












































