+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: PDB / ID: 9gu3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Human adult muscle nAChR in desensitised state in nanodisc with 1 mM acetylcholine | |||||||||
 要素 要素 |
| |||||||||
 キーワード キーワード | MEMBRANE PROTEIN / Ligand-gated ion channel / nicotinic receptor / pLGIC / cys-loop receptor | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報postsynaptic membrane organization / skeletal muscle tissue growth / musculoskeletal movement / Highly sodium permeable postsynaptic acetylcholine nicotinic receptors / Highly calcium permeable nicotinic acetylcholine receptors / Highly calcium permeable postsynaptic nicotinic acetylcholine receptors / acetylcholine receptor activity / acetylcholine-gated channel complex / behavioral response to nicotine / neuromuscular synaptic transmission ...postsynaptic membrane organization / skeletal muscle tissue growth / musculoskeletal movement / Highly sodium permeable postsynaptic acetylcholine nicotinic receptors / Highly calcium permeable nicotinic acetylcholine receptors / Highly calcium permeable postsynaptic nicotinic acetylcholine receptors / acetylcholine receptor activity / acetylcholine-gated channel complex / behavioral response to nicotine / neuromuscular synaptic transmission / acetylcholine-gated monoatomic cation-selective channel activity / muscle cell development / acetylcholine binding / synaptic transmission, cholinergic / monoatomic cation transmembrane transporter activity / nervous system process / acetylcholine receptor signaling pathway / postsynaptic specialization membrane / ligand-gated monoatomic ion channel activity / neuromuscular process / muscle cell cellular homeostasis / neuromuscular junction development / monoatomic cation transport / membrane depolarization / skeletal muscle contraction / neuronal action potential / muscle contraction / bioluminescence / generation of precursor metabolites and energy / regulation of membrane potential / response to nicotine / transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential / neuromuscular junction / neuron cellular homeostasis / transmembrane signaling receptor activity / channel activity / monoatomic ion transmembrane transport / chemical synaptic transmission / postsynaptic membrane / neuron projection / synapse / cell surface / signal transduction / plasma membrane 類似検索 - 分子機能 | |||||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト)  | |||||||||
| 手法 | 電子顕微鏡法 / 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 2.64 Å | |||||||||
 データ登録者 データ登録者 | Li, A. / Pike, A.C.W. / Chi, G. / Webster, R. / Maxwell, S. / Liu, W. / Beeson, D. / Sauer, D.B. / Dong, Y.Y. | |||||||||
| 資金援助 |  英国, 2件 英国, 2件
| |||||||||
 引用 引用 |  ジャーナル: Cell Rep / 年: 2025 ジャーナル: Cell Rep / 年: 2025タイトル: Structures of the human adult muscle-type nicotinic receptor in resting and desensitized states. 著者: Anna Li / Ashley C W Pike / Richard Webster / Susan Maxwell / Wei-Wei Liu / Gamma Chi / Jacqueline Palace / David Beeson / David B Sauer / Yin Yao Dong /  要旨: Muscle-type nicotinic acetylcholine receptor (AChR) is the key signaling molecule in neuromuscular junctions. Here, we present the structures of full-length human adult receptors in complex with ...Muscle-type nicotinic acetylcholine receptor (AChR) is the key signaling molecule in neuromuscular junctions. Here, we present the structures of full-length human adult receptors in complex with Fab35 in α-bungarotoxin (αBuTx)-bound resting states and ACh-bound desensitized states. In addition to identifying the conformational changes during recovery from desensitization, we also used electrophysiology to probe the effects of eight previously unstudied AChR genetic variants found in patients with congenital myasthenic syndrome (CMS), revealing they cause either slow- or fast-channel CMS characterized by prolonged or abbreviated ion channel bursts. The combined kinetic and structural data offer a better understanding of both the AChR state transition and the pathogenic mechanisms of disease variants. #1: ジャーナル: Acta Crystallogr D Struct Biol / 年: 2019 タイトル: Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. 著者: Dorothee Liebschner / Pavel V Afonine / Matthew L Baker / Gábor Bunkóczi / Vincent B Chen / Tristan I Croll / Bradley Hintze / Li Wei Hung / Swati Jain / Airlie J McCoy / Nigel W Moriarty / ...著者: Dorothee Liebschner / Pavel V Afonine / Matthew L Baker / Gábor Bunkóczi / Vincent B Chen / Tristan I Croll / Bradley Hintze / Li Wei Hung / Swati Jain / Airlie J McCoy / Nigel W Moriarty / Robert D Oeffner / Billy K Poon / Michael G Prisant / Randy J Read / Jane S Richardson / David C Richardson / Massimo D Sammito / Oleg V Sobolev / Duncan H Stockwell / Thomas C Terwilliger / Alexandre G Urzhumtsev / Lizbeth L Videau / Christopher J Williams / Paul D Adams /    要旨: Diffraction (X-ray, neutron and electron) and electron cryo-microscopy are powerful methods to determine three-dimensional macromolecular structures, which are required to understand biological ...Diffraction (X-ray, neutron and electron) and electron cryo-microscopy are powerful methods to determine three-dimensional macromolecular structures, which are required to understand biological processes and to develop new therapeutics against diseases. The overall structure-solution workflow is similar for these techniques, but nuances exist because the properties of the reduced experimental data are different. Software tools for structure determination should therefore be tailored for each method. Phenix is a comprehensive software package for macromolecular structure determination that handles data from any of these techniques. Tasks performed with Phenix include data-quality assessment, map improvement, model building, the validation/rebuilding/refinement cycle and deposition. Each tool caters to the type of experimental data. The design of Phenix emphasizes the automation of procedures, where possible, to minimize repetitive and time-consuming manual tasks, while default parameters are chosen to encourage best practice. A graphical user interface provides access to many command-line features of Phenix and streamlines the transition between programs, project tracking and re-running of previous tasks. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 構造ビューア | 分子:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- ダウンロードとリンク
ダウンロードとリンク
- ダウンロード
ダウンロード
| PDBx/mmCIF形式 |  9gu3.cif.gz 9gu3.cif.gz | 637.4 KB | 表示 |  PDBx/mmCIF形式 PDBx/mmCIF形式 |
|---|---|---|---|---|
| PDB形式 |  pdb9gu3.ent.gz pdb9gu3.ent.gz | 408.6 KB | 表示 |  PDB形式 PDB形式 |
| PDBx/mmJSON形式 |  9gu3.json.gz 9gu3.json.gz | ツリー表示 |  PDBx/mmJSON形式 PDBx/mmJSON形式 | |
| その他 |  その他のダウンロード その他のダウンロード |
-検証レポート
| 文書・要旨 |  9gu3_validation.pdf.gz 9gu3_validation.pdf.gz | 1.7 MB | 表示 |  wwPDB検証レポート wwPDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  9gu3_full_validation.pdf.gz 9gu3_full_validation.pdf.gz | 1.8 MB | 表示 | |
| XML形式データ |  9gu3_validation.xml.gz 9gu3_validation.xml.gz | 80.5 KB | 表示 | |
| CIF形式データ |  9gu3_validation.cif.gz 9gu3_validation.cif.gz | 123.3 KB | 表示 | |
| アーカイブディレクトリ |  https://data.pdbj.org/pub/pdb/validation_reports/gu/9gu3 https://data.pdbj.org/pub/pdb/validation_reports/gu/9gu3 ftp://data.pdbj.org/pub/pdb/validation_reports/gu/9gu3 ftp://data.pdbj.org/pub/pdb/validation_reports/gu/9gu3 | HTTPS FTP |
-関連構造データ
- リンク
リンク
- 集合体
集合体
| 登録構造単位 | 
|
|---|---|
| 1 |
|
- 要素
要素
-Acetylcholine receptor subunit ... , 4種, 5分子 ALBDE
| #1: タンパク質 | 分子量: 49747.645 Da / 分子数: 2 / 由来タイプ: 組換発現 / 由来: (組換発現)  Homo sapiens (ヒト) / 遺伝子: CHRNA1, ACHRA, CHNRA / プラスミド: TLCV2 / 細胞株 (発現宿主): Expi293F / 発現宿主: Homo sapiens (ヒト) / 遺伝子: CHRNA1, ACHRA, CHNRA / プラスミド: TLCV2 / 細胞株 (発現宿主): Expi293F / 発現宿主:  Homo sapiens (ヒト) / 参照: UniProt: P02708 Homo sapiens (ヒト) / 参照: UniProt: P02708#2: タンパク質 | | 分子量: 54596.477 Da / 分子数: 1 / 由来タイプ: 組換発現 / 由来: (組換発現)  Homo sapiens (ヒト) / 遺伝子: CHRNB1, ACHRB, CHRNB / プラスミド: TLCV2 / 細胞株 (発現宿主): Expi293F / 発現宿主: Homo sapiens (ヒト) / 遺伝子: CHRNB1, ACHRB, CHRNB / プラスミド: TLCV2 / 細胞株 (発現宿主): Expi293F / 発現宿主:  Homo sapiens (ヒト) / 参照: UniProt: P11230 Homo sapiens (ヒト) / 参照: UniProt: P11230#3: タンパク質 | | 分子量: 56915.359 Da / 分子数: 1 / 由来タイプ: 組換発現 / 由来: (組換発現)  Homo sapiens (ヒト) / 遺伝子: CHRND, ACHRD / プラスミド: TLCV2 / 細胞株 (発現宿主): Expi293F / 発現宿主: Homo sapiens (ヒト) / 遺伝子: CHRND, ACHRD / プラスミド: TLCV2 / 細胞株 (発現宿主): Expi293F / 発現宿主:  Homo sapiens (ヒト) / 参照: UniProt: Q07001 Homo sapiens (ヒト) / 参照: UniProt: Q07001#4: タンパク質 | | 分子量: 80603.750 Da / 分子数: 1 Mutation: EGFP insertion between residues R344 and A345 in the M3-M4 intracellular loop 由来タイプ: 組換発現 由来: (組換発現)  Homo sapiens (ヒト), (組換発現) Homo sapiens (ヒト), (組換発現)  遺伝子: CHRNE, ACHRE, GFP / プラスミド: TLCV2 / 細胞株 (発現宿主): Expi293F / 発現宿主:  Homo sapiens (ヒト) / 参照: UniProt: Q04844, UniProt: P42212 Homo sapiens (ヒト) / 参照: UniProt: Q04844, UniProt: P42212 |
|---|
-抗体 , 2種, 4分子 CGFH
| #5: 抗体 | 分子量: 23392.982 Da / 分子数: 2 / 由来タイプ: 組換発現 / 由来: (組換発現)   #6: 抗体 | 分子量: 23879.758 Da / 分子数: 2 / 由来タイプ: 組換発現 / 由来: (組換発現)   |
|---|
-糖 , 5種, 7分子 
| #7: 多糖 | | #8: 多糖 | #9: 多糖 | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose- ...2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | タイプ: oligosaccharide / 分子量: 1276.157 Da / 分子数: 1 / 由来タイプ: 組換発現 #10: 多糖 | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | #12: 糖 | ChemComp-NAG / | |
|---|
-非ポリマー , 1種, 2分子 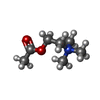
| #11: 化合物 |
|---|
-詳細
| 研究の焦点であるリガンドがあるか | Y |
|---|---|
| Has protein modification | Y |
-実験情報
-実験
| 実験 | 手法: 電子顕微鏡法 |
|---|---|
| EM実験 | 試料の集合状態: PARTICLE / 3次元再構成法: 単粒子再構成法 |
- 試料調製
試料調製
| 構成要素 | 名称: Human adult muscle nAChR in desensitised state in nanodisc with 1 mM acetylcholine タイプ: COMPLEX 詳細: nAChR in complex with acetylcholine and Fab35 in MSP2N2-soy polar lipids nanodiscs Entity ID: #1-#6 / 由来: RECOMBINANT | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 分子量 | 値: 0.386 MDa / 実験値: NO | |||||||||||||||
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) | |||||||||||||||
| 由来(組換発現) | 生物種:  Homo sapiens (ヒト) / 細胞: Expi293F / プラスミド: TLCV2 Homo sapiens (ヒト) / 細胞: Expi293F / プラスミド: TLCV2 | |||||||||||||||
| 緩衝液 | pH: 7.5 / 詳細: 20 mM HEPES pH 7.5, 150 mM NaCl | |||||||||||||||
| 緩衝液成分 |
| |||||||||||||||
| 試料 | 濃度: 2 mg/ml / 包埋: NO / シャドウイング: NO / 染色: NO / 凍結: YES / 詳細: This sample was monodisperse | |||||||||||||||
| 試料支持 | グリッドの材料: COPPER / グリッドのサイズ: 200 divisions/in. / グリッドのタイプ: Quantifoil R1.2/1.3 | |||||||||||||||
| 急速凍結 | 装置: FEI VITROBOT MARK IV / 凍結剤: ETHANE / 湿度: 100 % / 凍結前の試料温度: 277.35 K / 詳細: 2.5 ul sample, blot time=4 s, blot force=-10 |
- 電子顕微鏡撮影
電子顕微鏡撮影
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
|---|---|
| 顕微鏡 | モデル: TFS KRIOS |
| 電子銃 | 電子線源:  FIELD EMISSION GUN / 加速電圧: 300 kV / 照射モード: FLOOD BEAM FIELD EMISSION GUN / 加速電圧: 300 kV / 照射モード: FLOOD BEAM |
| 電子レンズ | モード: BRIGHT FIELD / 倍率(公称値): 130000 X / 最大 デフォーカス(公称値): 2200 nm / 最小 デフォーカス(公称値): 1000 nm / Cs: 2.7 mm / C2レンズ絞り径: 50 µm / アライメント法: COMA FREE |
| 試料ホルダ | 凍結剤: NITROGEN 試料ホルダーモデル: FEI TITAN KRIOS AUTOGRID HOLDER |
| 撮影 | 平均露光時間: 1.7 sec. / 電子線照射量: 50 e/Å2 フィルム・検出器のモデル: GATAN K3 BIOQUANTUM (6k x 4k) 撮影したグリッド数: 1 / 実像数: 19925 |
| 電子光学装置 | エネルギーフィルター名称: GIF Bioquantum / エネルギーフィルタースリット幅: 20 eV |
| 画像スキャン | 横: 4092 / 縦: 5760 |
- 解析
解析
| EMソフトウェア |
| ||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF補正 | タイプ: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||||||||||||||||
| 粒子像の選択 | 選択した粒子像数: 5168840 / 詳細: extracted by topaz picking | ||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 点対称性: C1 (非対称) | ||||||||||||||||||||||||||||||||||||||||||||
| 3次元再構成 | 解像度: 2.64 Å / 解像度の算出法: FSC 0.143 CUT-OFF / 粒子像の数: 434976 / アルゴリズム: FOURIER SPACE / 詳細: non-uniform refinenement / クラス平均像の数: 1 / 対称性のタイプ: POINT | ||||||||||||||||||||||||||||||||||||||||||||
| 原子モデル構築 | プロトコル: FLEXIBLE FIT / 空間: REAL 詳細: Initial fitting of alphafold models using chimeraX followed by manual rebuilding in COOT and final refinement in ISOLDE and PHENIX | ||||||||||||||||||||||||||||||||||||||||||||
| 原子モデル構築 |
| ||||||||||||||||||||||||||||||||||||||||||||
| 精密化 | 交差検証法: NONE 立体化学のターゲット値: GeoStd + Monomer Library + CDL v1.2 | ||||||||||||||||||||||||||||||||||||||||||||
| 原子変位パラメータ | Biso mean: 159.81 Å2 | ||||||||||||||||||||||||||||||||||||||||||||
| 拘束条件 |
|
 ムービー
ムービー コントローラー
コントローラー










 PDBj
PDBj







