[English] 日本語
 Yorodumi
Yorodumi- PDB-9der: Cryo-EM Structures of Full-Length Integrin alphaIIbbeta3 in Nativ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 9der | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structures of Full-Length Integrin alphaIIbbeta3 in Native Lipids Complexed with Tirofiban | ||||||||||||
 Components Components |
| ||||||||||||
 Keywords Keywords | CELL ADHESION / complex / open headpiece | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of serotonin uptake / positive regulation of adenylate cyclase-inhibiting opioid receptor signaling pathway / tube development / alpha9-beta1 integrin-ADAM8 complex / regulation of trophoblast cell migration / integrin alphaIIb-beta3 complex / regulation of postsynaptic neurotransmitter receptor diffusion trapping / alphav-beta3 integrin-vitronectin complex / maintenance of postsynaptic specialization structure / regulation of extracellular matrix organization ...regulation of serotonin uptake / positive regulation of adenylate cyclase-inhibiting opioid receptor signaling pathway / tube development / alpha9-beta1 integrin-ADAM8 complex / regulation of trophoblast cell migration / integrin alphaIIb-beta3 complex / regulation of postsynaptic neurotransmitter receptor diffusion trapping / alphav-beta3 integrin-vitronectin complex / maintenance of postsynaptic specialization structure / regulation of extracellular matrix organization / positive regulation of glomerular mesangial cell proliferation / platelet alpha granule membrane / integrin alphav-beta3 complex / negative regulation of lipoprotein metabolic process / alphav-beta3 integrin-PKCalpha complex / fibrinogen binding / alphav-beta3 integrin-HMGB1 complex / vascular endothelial growth factor receptor 2 binding / negative regulation of lipid transport / positive regulation of vascular endothelial growth factor signaling pathway / Elastic fibre formation / mesodermal cell differentiation / cell-substrate junction assembly / alphav-beta3 integrin-IGF-1-IGF1R complex / positive regulation of bone resorption / platelet-derived growth factor receptor binding / glycinergic synapse / regulation of release of sequestered calcium ion into cytosol / filopodium membrane / extracellular matrix binding / wound healing, spreading of epidermal cells / positive regulation of vascular endothelial growth factor receptor signaling pathway / apolipoprotein A-I-mediated signaling pathway / positive regulation of cell adhesion mediated by integrin / positive regulation of leukocyte migration / negative regulation of low-density lipoprotein particle clearance / regulation of bone resorption / angiogenesis involved in wound healing / apoptotic cell clearance / positive regulation of fibroblast migration / integrin complex / positive regulation of smooth muscle cell migration / heterotypic cell-cell adhesion / smooth muscle cell migration / Molecules associated with elastic fibres / cell adhesion mediated by integrin / negative chemotaxis / positive regulation of cell-matrix adhesion / Mechanical load activates signaling by PIEZO1 and integrins in osteocytes / Syndecan interactions / p130Cas linkage to MAPK signaling for integrins / cellular response to insulin-like growth factor stimulus / protein disulfide isomerase activity / regulation of postsynaptic neurotransmitter receptor internalization / positive regulation of osteoblast proliferation / microvillus membrane / cell-substrate adhesion / platelet-derived growth factor receptor signaling pathway / PECAM1 interactions / GRB2:SOS provides linkage to MAPK signaling for Integrins / TGF-beta receptor signaling activates SMADs / fibronectin binding / lamellipodium membrane / blood coagulation, fibrin clot formation / negative regulation of macrophage derived foam cell differentiation / negative regulation of lipid storage / ECM proteoglycans / Integrin cell surface interactions / negative regulation of endothelial cell apoptotic process / positive regulation of T cell migration / coreceptor activity / cellular response to platelet-derived growth factor stimulus / positive regulation of endothelial cell proliferation / Integrin signaling / cell adhesion molecule binding / positive regulation of substrate adhesion-dependent cell spreading / positive regulation of endothelial cell migration / positive regulation of smooth muscle cell proliferation / substrate adhesion-dependent cell spreading / embryo implantation / protein kinase C binding / Turbulent (oscillatory, disturbed) flow shear stress activates signaling by PIEZO1 and integrins in endothelial cells / cell-matrix adhesion / response to activity / integrin-mediated signaling pathway / Signal transduction by L1 / regulation of actin cytoskeleton organization / wound healing / cellular response to mechanical stimulus / cell-cell adhesion / Signaling by high-kinase activity BRAF mutants / RUNX1 regulates genes involved in megakaryocyte differentiation and platelet function / MAP2K and MAPK activation / platelet activation / VEGFA-VEGFR2 Pathway / platelet aggregation / integrin binding / cellular response to xenobiotic stimulus / ruffle membrane / positive regulation of fibroblast proliferation Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.9 Å | ||||||||||||
 Authors Authors | Adair, B. / Xiong, J.P. / Yeager, M. / Arnaout, M.A. | ||||||||||||
| Funding support |  United States, 3items United States, 3items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Platelet integrin αIIbβ3 plays a key role in a venous thrombogenesis mouse model. Authors: Brian D Adair / Conroy O Field / José L Alonso / Jian-Ping Xiong / Shi-Xian Deng / Hyun Sook Ahn / Eivgeni Mashin / Clary B Clish / Johannes van Agthoven / Mark Yeager / Youzhong Guo / ...Authors: Brian D Adair / Conroy O Field / José L Alonso / Jian-Ping Xiong / Shi-Xian Deng / Hyun Sook Ahn / Eivgeni Mashin / Clary B Clish / Johannes van Agthoven / Mark Yeager / Youzhong Guo / David A Tess / Donald W Landry / Mortimer Poncz / M Amin Arnaout /  Abstract: Venous thrombosis (VT) is a common vascular disease associated with reduced survival and a high recurrence rate. VT is initiated by the accumulation of platelets and neutrophils at sites of ...Venous thrombosis (VT) is a common vascular disease associated with reduced survival and a high recurrence rate. VT is initiated by the accumulation of platelets and neutrophils at sites of endothelial cell activation. A role for platelet αIIbβ3 in VT is not established, a task complicated by the increased bleeding risk caused by partial agonists such as tirofiban. Here, we show that m-tirofiban, a modified version of tirofiban, does not agonize αIIbβ3 based on lack of neoepitope expression and the cryo-EM structure of m-tirofiban/full-length αIIbβ3 complex. m-tirofiban abolishes agonist-induced platelet aggregation while preserving clot retraction ex vivo and, unlike tirofiban, it suppresses venous thrombogenesis in a mouse model without increasing bleeding. These findings establish a key role for αIIbβ3 in VT initiation and suggest that m-tirofiban and compounds with a similar structurally-defined mechanism of action merit consideration as potential thromboprophylaxis agents in patients at high risk for VT and hemorrhage. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  9der.cif.gz 9der.cif.gz | 334 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb9der.ent.gz pdb9der.ent.gz | 261.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  9der.json.gz 9der.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/de/9der https://data.pdbj.org/pub/pdb/validation_reports/de/9der ftp://data.pdbj.org/pub/pdb/validation_reports/de/9der ftp://data.pdbj.org/pub/pdb/validation_reports/de/9der | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  46794MC  9deqC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Protein , 2 types, 2 molecules AB
| #1: Protein | Mass: 110106.391 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) / References: UniProt: P08514 Homo sapiens (human) / References: UniProt: P08514 |
|---|---|
| #2: Protein | Mass: 84478.500 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) / References: UniProt: P05106 Homo sapiens (human) / References: UniProt: P05106 |
-Sugars , 4 types, 5 molecules
| #3: Polysaccharide | Source method: isolated from a genetically manipulated source #4: Polysaccharide | beta-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4) ...beta-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | Type: oligosaccharide / Mass: 748.682 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source #5: Polysaccharide | beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4) ...beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | Type: oligosaccharide / Mass: 748.682 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source #6: Polysaccharide | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | Source method: isolated from a genetically manipulated source |
|---|
-Non-polymers , 4 types, 10 molecules 

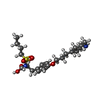




| #7: Chemical | ChemComp-CA / #8: Chemical | ChemComp-MG / | #9: Chemical | ChemComp-AGG / | #10: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: CELL / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: full-length integrin alphaIIbbeta3 / Type: COMPLEX / Entity ID: #1-#2 / Source: NATURAL |
|---|---|
| Molecular weight | Experimental value: NO |
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Buffer solution | pH: 7.4 / Details: TBS/1mM CaCl2 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 81000 X / Nominal defocus max: 2500 nm / Nominal defocus min: 1000 nm / Cs: 0.27 mm |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Electron dose: 50 e/Å2 / Film or detector model: GATAN K3 (6k x 4k) / Num. of real images: 11947 |
| EM imaging optics | Energyfilter name: GIF Quantum ER / Energyfilter slit width: 10 eV |
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 4245069 | ||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3.9 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 97969 / Symmetry type: POINT | ||||||||||||||||||||||||||||||||
| Atomic model building | Protocol: FLEXIBLE FIT / Space: REAL | ||||||||||||||||||||||||||||||||
| Atomic model building | PDB-ID: 8T2U Accession code: 8T2U / Source name: PDB / Type: experimental model | ||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller



 PDBj
PDBj















