+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 9c1f | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mink RyR3 in open conformation bound to Ca2+/ATP/caffeine | ||||||||||||
 Components Components |
| ||||||||||||
 Keywords Keywords | MEMBRANE PROTEIN / RyR3 / Ryanodine Receptor / calcium channel / ion channel | ||||||||||||
| Function / homology |  Function and homology information Function and homology information: / negative regulation of calcium-mediated signaling / negative regulation of insulin secretion involved in cellular response to glucose stimulus / neuronal action potential propagation / negative regulation of release of sequestered calcium ion into cytosol / insulin secretion involved in cellular response to glucose stimulus / ryanodine-sensitive calcium-release channel activity / release of sequestered calcium ion into cytosol by sarcoplasmic reticulum / response to redox state / negative regulation of heart rate ...: / negative regulation of calcium-mediated signaling / negative regulation of insulin secretion involved in cellular response to glucose stimulus / neuronal action potential propagation / negative regulation of release of sequestered calcium ion into cytosol / insulin secretion involved in cellular response to glucose stimulus / ryanodine-sensitive calcium-release channel activity / release of sequestered calcium ion into cytosol by sarcoplasmic reticulum / response to redox state / negative regulation of heart rate / 'de novo' protein folding / FK506 binding / smooth endoplasmic reticulum / smooth muscle contraction / T cell proliferation / striated muscle contraction / regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion / calcium channel inhibitor activity / regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum / Ion homeostasis / release of sequestered calcium ion into cytosol / calcium channel complex / sarcoplasmic reticulum membrane / protein maturation / peptidylprolyl isomerase / calcium channel regulator activity / peptidyl-prolyl cis-trans isomerase activity / calcium-mediated signaling / sarcolemma / Stimuli-sensing channels / Z disc / intracellular calcium ion homeostasis / positive regulation of cytosolic calcium ion concentration / protein refolding / transmembrane transporter binding / calmodulin binding / signaling receptor binding / calcium ion binding / membrane / cytoplasm Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) Neogale vison (American mink) Neogale vison (American mink) | ||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.22 Å | ||||||||||||
 Authors Authors | Chen, Y.S. / Van Petegem, F. | ||||||||||||
| Funding support |  Canada, 3items Canada, 3items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Cryo-EM investigation of ryanodine receptor type 3. Authors: Yu Seby Chen / Maricela Garcia-Castañeda / Maria Charalambous / Daniela Rossi / Vincenzo Sorrentino / Filip Van Petegem /   Abstract: Ryanodine Receptor isoform 3 (RyR3) is a large ion channel found in the endoplasmic reticulum membrane of many different cell types. Within the hippocampal region of the brain, it is found in ...Ryanodine Receptor isoform 3 (RyR3) is a large ion channel found in the endoplasmic reticulum membrane of many different cell types. Within the hippocampal region of the brain, it is found in dendritic spines and regulates synaptic plasticity. It controls myogenic tone in arteries and is upregulated in skeletal muscle in early development. RyR3 has a unique functional profile with a very high sensitivity to activating ligands, enabling high gain in Ca-induced Ca release. Here we solve high-resolution cryo-EM structures of RyR3 in non-activating and activating conditions, revealing structural transitions that occur during channel opening. Addition of activating ligands yields only open channels, indicating an intrinsically high open probability under these conditions. RyR3 has reduced binding affinity to the auxiliary protein FKBP12.6 due to several sequence variations in the binding interface. We map disease-associated sequence variants and binding sites for known pharmacological agents. The N-terminal region contains ligand binding sites for a putative chloride anion and ATP, both of which are targeted by sequence variants linked to epileptic encephalopathy. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  9c1f.cif.gz 9c1f.cif.gz | 3.2 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb9c1f.ent.gz pdb9c1f.ent.gz | Display |  PDB format PDB format | |
| PDBx/mmJSON format |  9c1f.json.gz 9c1f.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/c1/9c1f https://data.pdbj.org/pub/pdb/validation_reports/c1/9c1f ftp://data.pdbj.org/pub/pdb/validation_reports/c1/9c1f ftp://data.pdbj.org/pub/pdb/validation_reports/c1/9c1f | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  45117MC  9c1eC C: citing same article ( M: map data used to model this data |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Protein , 2 types, 8 molecules ACEGBDFH
| #1: Protein | Mass: 11939.562 Da / Num. of mol.: 4 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: FKBP1B, FKBP12.6, FKBP1L, FKBP9, OTK4 / Production host: Homo sapiens (human) / Gene: FKBP1B, FKBP12.6, FKBP1L, FKBP9, OTK4 / Production host:  #2: Protein | Mass: 551217.375 Da / Num. of mol.: 4 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Neogale vison (American mink) / Gene: RYR3 / Cell line (production host): HEK293T / Production host: Neogale vison (American mink) / Gene: RYR3 / Cell line (production host): HEK293T / Production host:  Homo sapiens (human) / References: UniProt: A0A8C7B9Y8 Homo sapiens (human) / References: UniProt: A0A8C7B9Y8 |
|---|
-Non-polymers , 5 types, 24 molecules 


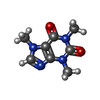





| #3: Chemical | ChemComp-ZN / #4: Chemical | ChemComp-CA / #5: Chemical | ChemComp-ATP / #6: Chemical | ChemComp-CFF / #7: Chemical | ChemComp-CL / |
|---|
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: mink RyR3 + FKBP12.6 / Type: COMPLEX / Entity ID: #1-#2 / Source: RECOMBINANT |
|---|---|
| Molecular weight | Value: 2.25 MDa / Experimental value: YES |
| Source (natural) | Organism:  Neogale vison (American mink) Neogale vison (American mink) |
| Source (recombinant) | Organism:  Homo sapiens (human) / Cell: HEK293T Homo sapiens (human) / Cell: HEK293T |
| Buffer solution | pH: 7.5 Details: 20 mM HEPES pH 7.5, 250 mM NaCl, 2 mM DTT, 0.02% GDN, 0.03 mM free Ca2+ buffered with 2 mM EGTA, 5 mM ATP, 5 mM caffeine, 1:1000 diluted protease inhibitor cocktail |
| Specimen | Conc.: 5 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Specimen support | Grid material: GOLD / Grid mesh size: 300 divisions/in. / Grid type: UltrAuFoil R1.2/1.3 |
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 277.15 K Details: blotted for 3 s (blot force of 7 to 10) using ashless blotting paper (Whatman) |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal defocus max: 2000 nm / Nominal defocus min: 500 nm / Cs: 2.7 mm |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Electron dose: 50 e/Å2 / Film or detector model: FEI FALCON IV (4k x 4k) / Num. of grids imaged: 3 / Num. of real images: 10099 |
| EM imaging optics | Energyfilter name: TFS Selectris |
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 162679 | ||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Point symmetry: C4 (4 fold cyclic) | ||||||||||||||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3.22 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 84690 / Algorithm: FOURIER SPACE / Num. of class averages: 1 / Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||||||||||
| Atomic model building | Protocol: RIGID BODY FIT / Space: REAL / Target criteria: cross-correlation coefficient | ||||||||||||||||||||||||||||||||||||||||||||
| Atomic model building | PDB-ID: 9C1E Accession code: 9C1E / Details: mink RyR3 in closed conformation / Source name: PDB / Type: experimental model |
 Movie
Movie Controller
Controller














 PDBj
PDBj









