| Entry | Database: PDB / ID: 9bpx
|
|---|
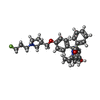
| Title | Estrogen Receptor Alpha Ligand Binding Domain Y537S Mutant in Complex with (1'-(4-((1-butylpyrrolidin-3-yl)methoxy)phenyl)-6'-hydroxy-1',4'-dihydro-2'H-spiro[cyclopropane-1,3'-isoquinolin]-2'-yl)(phenyl)methanone |
|---|
 Components Components | Estrogen receptor |
|---|
 Keywords Keywords | TRANSCRIPTION / Estrogen / Alpha Helical Bundle / Breast Cancer |
|---|
| Function / homology |  Function and homology information Function and homology information
regulation of epithelial cell apoptotic process / antral ovarian follicle growth / regulation of branching involved in prostate gland morphogenesis / RUNX1 regulates transcription of genes involved in WNT signaling / RUNX1 regulates estrogen receptor mediated transcription / regulation of toll-like receptor signaling pathway / nuclear estrogen receptor activity / epithelial cell development / steroid hormone receptor signaling pathway / prostate epithelial cord elongation ...regulation of epithelial cell apoptotic process / antral ovarian follicle growth / regulation of branching involved in prostate gland morphogenesis / RUNX1 regulates transcription of genes involved in WNT signaling / RUNX1 regulates estrogen receptor mediated transcription / regulation of toll-like receptor signaling pathway / nuclear estrogen receptor activity / epithelial cell development / steroid hormone receptor signaling pathway / prostate epithelial cord elongation / epithelial cell proliferation involved in mammary gland duct elongation / prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis / mammary gland branching involved in pregnancy / uterus development / negative regulation of smooth muscle cell apoptotic process / vagina development / TFIIB-class transcription factor binding / androgen metabolic process / mammary gland alveolus development / cellular response to estrogen stimulus / estrogen response element binding / Mitochondrial unfolded protein response (UPRmt) / nuclear receptor-mediated steroid hormone signaling pathway / Nuclear signaling by ERBB4 / : / : / RNA polymerase II preinitiation complex assembly / positive regulation of nitric-oxide synthase activity / estrogen receptor signaling pathway / protein localization to chromatin / steroid binding / 14-3-3 protein binding / TFAP2 (AP-2) family regulates transcription of growth factors and their receptors / negative regulation of canonical NF-kappaB signal transduction / ESR-mediated signaling / negative regulation of miRNA transcription / TBP-class protein binding / nitric-oxide synthase regulator activity / nuclear estrogen receptor binding / transcription corepressor binding / transcription coregulator binding / stem cell differentiation / SUMOylation of intracellular receptors / cellular response to estradiol stimulus / euchromatin / beta-catenin binding / Nuclear Receptor transcription pathway / response to estrogen / transcription coactivator binding / male gonad development / nuclear receptor activity / positive regulation of fibroblast proliferation / Constitutive Signaling by Aberrant PI3K in Cancer / sequence-specific double-stranded DNA binding / positive regulation of nitric oxide biosynthetic process / Regulation of RUNX2 expression and activity / Ovarian tumor domain proteases / response to estradiol / PIP3 activates AKT signaling / positive regulation of cytosolic calcium ion concentration / ATPase binding / PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling / regulation of inflammatory response / DNA-binding transcription activator activity, RNA polymerase II-specific / fibroblast proliferation / transcription regulator complex / phospholipase C-activating G protein-coupled receptor signaling pathway / Estrogen-dependent gene expression / DNA-binding transcription factor activity, RNA polymerase II-specific / calmodulin binding / Extra-nuclear estrogen signaling / RNA polymerase II cis-regulatory region sequence-specific DNA binding / chromatin remodeling / DNA-binding transcription factor activity / negative regulation of gene expression / chromatin binding / regulation of DNA-templated transcription / regulation of transcription by RNA polymerase II / protein kinase binding / chromatin / positive regulation of DNA-templated transcription / enzyme binding / negative regulation of transcription by RNA polymerase II / Golgi apparatus / signal transduction / positive regulation of transcription by RNA polymerase II / protein-containing complex / zinc ion binding / nucleoplasm / identical protein binding / nucleus / membrane / plasma membrane / cytosol / cytoplasmSimilarity search - Function Oestrogen-type nuclear receptor final C-terminal domain / Oestrogen-type nuclear receptor final C-terminal / Estrogen receptor / : / Oestrogen receptor / Estrogen receptor/oestrogen-related receptor / : / Nuclear hormone receptor / Nuclear hormones receptors DNA-binding region signature. / Zinc finger, nuclear hormone receptor-type ...Oestrogen-type nuclear receptor final C-terminal domain / Oestrogen-type nuclear receptor final C-terminal / Estrogen receptor / : / Oestrogen receptor / Estrogen receptor/oestrogen-related receptor / : / Nuclear hormone receptor / Nuclear hormones receptors DNA-binding region signature. / Zinc finger, nuclear hormone receptor-type / Double treble clef zinc finger, C4 type / Nuclear hormone receptors DNA-binding domain profile. / c4 zinc finger in nuclear hormone receptors / Nuclear hormone receptor, ligand-binding domain / Nuclear hormone receptor-like domain superfamily / Ligand-binding domain of nuclear hormone receptor / Nuclear receptor (NR) ligand-binding (LBD) domain profile. / Ligand binding domain of hormone receptors / Zinc finger, NHR/GATA-typeSimilarity search - Domain/homology |
|---|
| Biological species |  Homo sapiens (human) Homo sapiens (human) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.2 Å MOLECULAR REPLACEMENT / Resolution: 2.2 Å |
|---|
 Authors Authors | Young, K.Y. / Fanning, S.W. |
|---|
| Funding support |  United States, 1items United States, 1items | Organization | Grant number | Country |
|---|
| National Institutes of Health/National Cancer Institute (NIH/NCI) | 5R37CA279341 |  United States United States |
|
|---|
 Citation Citation |  Journal: Breast Cancer Res / Year: 2025 Journal: Breast Cancer Res / Year: 2025
Title: Targeting unique ligand binding domain structural features downregulates DKK1 in Y537S ESR1 mutant breast cancer cells.
Authors: Young, K.S. / Hancock, G.R. / Fink, E.C. / Zigrossi, A. / Flowers, B. / Cooper, D.A. / Nguyen, V.T. / Martinez, M.C. / Mon, K.S. / Bosland, M. / Zak, D.R. / Runde, A.P. / Sharifi, M.N. / ...Authors: Young, K.S. / Hancock, G.R. / Fink, E.C. / Zigrossi, A. / Flowers, B. / Cooper, D.A. / Nguyen, V.T. / Martinez, M.C. / Mon, K.S. / Bosland, M. / Zak, D.R. / Runde, A.P. / Sharifi, M.N. / Kastrati, I. / Minh, D.D.L. / Kregel, S. / Fanning, S.W. |
|---|
| History | | Deposition | May 8, 2024 | Deposition site: RCSB / Processing site: RCSB |
|---|
| Revision 1.0 | Feb 12, 2025 | Provider: repository / Type: Initial release |
|---|
|
|---|
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Homo sapiens (human)
Homo sapiens (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.2 Å
MOLECULAR REPLACEMENT / Resolution: 2.2 Å  Authors
Authors United States, 1items
United States, 1items  Citation
Citation Journal: Breast Cancer Res / Year: 2025
Journal: Breast Cancer Res / Year: 2025 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 9bpx.cif.gz
9bpx.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb9bpx.ent.gz
pdb9bpx.ent.gz PDB format
PDB format 9bpx.json.gz
9bpx.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads 9bpx_validation.pdf.gz
9bpx_validation.pdf.gz wwPDB validaton report
wwPDB validaton report 9bpx_full_validation.pdf.gz
9bpx_full_validation.pdf.gz 9bpx_validation.xml.gz
9bpx_validation.xml.gz 9bpx_validation.cif.gz
9bpx_validation.cif.gz https://data.pdbj.org/pub/pdb/validation_reports/bp/9bpx
https://data.pdbj.org/pub/pdb/validation_reports/bp/9bpx ftp://data.pdbj.org/pub/pdb/validation_reports/bp/9bpx
ftp://data.pdbj.org/pub/pdb/validation_reports/bp/9bpx

 F&H Search
F&H Search Links
Links Assembly
Assembly
 Components
Components Homo sapiens (human) / Gene: ESR1, ESR, NR3A1 / Production host:
Homo sapiens (human) / Gene: ESR1, ESR, NR3A1 / Production host: 
 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  NSLS-II
NSLS-II  / Beamline: 17-ID-1 / Wavelength: 1 Å
/ Beamline: 17-ID-1 / Wavelength: 1 Å Processing
Processing MOLECULAR REPLACEMENT / Resolution: 2.2→45.63 Å / SU ML: 0.22 / Cross valid method: FREE R-VALUE / σ(F): 1.36 / Phase error: 21.98 / Stereochemistry target values: ML
MOLECULAR REPLACEMENT / Resolution: 2.2→45.63 Å / SU ML: 0.22 / Cross valid method: FREE R-VALUE / σ(F): 1.36 / Phase error: 21.98 / Stereochemistry target values: ML Movie
Movie Controller
Controller


 PDBj
PDBj