[English] 日本語
 Yorodumi
Yorodumi- PDB-9bim: Cryo-EM structure of human CHT1 in the HC-3 bound outward-facing state -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 9bim | |||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human CHT1 in the HC-3 bound outward-facing state | |||||||||||||||||||||||||||||||||||||||
 Components Components | High affinity choline transporter 1 | |||||||||||||||||||||||||||||||||||||||
 Keywords Keywords | TRANSPORT PROTEIN / Choline transporter | |||||||||||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationcholine:sodium symporter activity / Defective SLC5A7 causes distal hereditary motor neuronopathy 7A (HMN7A) / Defective SLC5A7 causes distal hereditary motor neuronopathy 7A (HMN7A) / acetylcholine biosynthetic process / : / choline transmembrane transporter activity / Acetylcholine Neurotransmitter Release Cycle / choline transport / neuromuscular synaptic transmission / choline binding ...choline:sodium symporter activity / Defective SLC5A7 causes distal hereditary motor neuronopathy 7A (HMN7A) / Defective SLC5A7 causes distal hereditary motor neuronopathy 7A (HMN7A) / acetylcholine biosynthetic process / : / choline transmembrane transporter activity / Acetylcholine Neurotransmitter Release Cycle / choline transport / neuromuscular synaptic transmission / choline binding / synaptic transmission, cholinergic / neurotransmitter transport / neuromuscular junction / transmembrane transport / synaptic vesicle membrane / presynaptic membrane / early endosome membrane / perikaryon / in utero embryonic development / axon / synapse / dendrite / membrane / plasma membrane Similarity search - Function | |||||||||||||||||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||||||||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.67 Å | |||||||||||||||||||||||||||||||||||||||
 Authors Authors | Xue, J. / Jiang, Y. | |||||||||||||||||||||||||||||||||||||||
| Funding support |  United States, 3items United States, 3items
| |||||||||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Cell Discov / Year: 2024 Journal: Cell Discov / Year: 2024Title: Structural mechanisms of human sodium-coupled high-affinity choline transporter CHT1. Authors: Jing Xue / Hongwen Chen / Yong Wang / Youxing Jiang /   Abstract: Mammalian sodium-coupled high-affinity choline transporter CHT1 uptakes choline in cholinergic neurons for acetylcholine synthesis and plays a critical role in cholinergic neurotransmission. Here, we ...Mammalian sodium-coupled high-affinity choline transporter CHT1 uptakes choline in cholinergic neurons for acetylcholine synthesis and plays a critical role in cholinergic neurotransmission. Here, we present the high-resolution cryo-EM structures of human CHT1 in apo, substrate- and ion-bound, hemicholinium-3-inhibited, and ML352-inhibited states. These structures represent three distinct conformational states, elucidating the structural basis of the CHT1-mediated choline uptake mechanism. Three ion-binding sites, two for Na and one for Cl, are unambiguously defined in the structures, demonstrating that both ions are indispensable cofactors for high-affinity choline-binding and are likely transported together with the substrate in a 2:1:1 stoichiometry. The two inhibitor-bound CHT1 structures reveal two distinct inhibitory mechanisms and provide a potential structural platform for designing therapeutic drugs to manipulate cholinergic neuron activity. Combined with the functional analysis, this study provides a comprehensive view of the structural mechanisms underlying substrate specificity, substrate/ion co-transport, and drug inhibition of a physiologically important symporter. | |||||||||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  9bim.cif.gz 9bim.cif.gz | 111.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb9bim.ent.gz pdb9bim.ent.gz | 82.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  9bim.json.gz 9bim.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  9bim_validation.pdf.gz 9bim_validation.pdf.gz | 1.3 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  9bim_full_validation.pdf.gz 9bim_full_validation.pdf.gz | 1.3 MB | Display | |
| Data in XML |  9bim_validation.xml.gz 9bim_validation.xml.gz | 30.1 KB | Display | |
| Data in CIF |  9bim_validation.cif.gz 9bim_validation.cif.gz | 42.8 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/bi/9bim https://data.pdbj.org/pub/pdb/validation_reports/bi/9bim ftp://data.pdbj.org/pub/pdb/validation_reports/bi/9bim ftp://data.pdbj.org/pub/pdb/validation_reports/bi/9bim | HTTPS FTP |
-Related structure data
| Related structure data |  44593MC  9bfiC  9bfjC  9bfkC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein | Mass: 66526.625 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: SLC5A7, CHT1 / Production host: Homo sapiens (human) / Gene: SLC5A7, CHT1 / Production host:  Homo sapiens (human) / References: UniProt: Q9GZV3 Homo sapiens (human) / References: UniProt: Q9GZV3 |
|---|---|
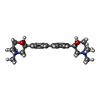
| #2: Chemical | ChemComp-HC6 / ( |
| Has ligand of interest | Y |
| Has protein modification | N |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: human CHT1 in the HC-3 bound state / Type: ORGANELLE OR CELLULAR COMPONENT / Entity ID: #1 / Source: RECOMBINANT |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Source (recombinant) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Buffer solution | pH: 7.4 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal defocus max: 1800 nm / Nominal defocus min: 800 nm |
| Image recording | Electron dose: 60 e/Å2 / Film or detector model: FEI FALCON IV (4k x 4k) |
- Processing
Processing
| EM software | Name: PHENIX / Category: model refinement | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3.67 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 191427 / Symmetry type: POINT | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller





 PDBj
PDBj