+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8wwy | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of mouse TIFA/TIFAB heterodimer | ||||||
 Components Components |
| ||||||
 Keywords Keywords | SIGNALING PROTEIN / Complex | ||||||
| Function / homology |  Function and homology information Function and homology informationregulation of muscle organ development / learned vocalization behavior / negative regulation of saliva secretion / hard palate morphogenesis / negative regulation of relaxation of muscle / trigeminal nerve development / regulation of intrinsic apoptotic signaling pathway in response to osmotic stress by p53 class mediator / craniofacial suture morphogenesis / thorax and anterior abdomen determination / vestibulocochlear nerve formation ...regulation of muscle organ development / learned vocalization behavior / negative regulation of saliva secretion / hard palate morphogenesis / negative regulation of relaxation of muscle / trigeminal nerve development / regulation of intrinsic apoptotic signaling pathway in response to osmotic stress by p53 class mediator / craniofacial suture morphogenesis / thorax and anterior abdomen determination / vestibulocochlear nerve formation / Alpha-protein kinase 1 signaling pathway / genitalia morphogenesis / TAK1-dependent IKK and NF-kappa-B activation / mastication / peristalsis / cochlea morphogenesis / auditory behavior / myeloid cell differentiation / deubiquitinase activator activity / neuromuscular process controlling balance / toll-like receptor signaling pathway / cytoplasmic pattern recognition receptor signaling pathway / hematopoietic progenitor cell differentiation / lipopolysaccharide-mediated signaling pathway / tumor necrosis factor-mediated signaling pathway / protein homooligomerization / regulation of gene expression / positive regulation of canonical NF-kappaB signal transduction / innate immune response / cytoplasm Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.79 Å MOLECULAR REPLACEMENT / Resolution: 1.79 Å | ||||||
 Authors Authors | Nakamura, T. | ||||||
| Funding support |  Japan, 1items Japan, 1items
| ||||||
 Citation Citation |  Journal: Proc.Natl.Acad.Sci.USA / Year: 2024 Journal: Proc.Natl.Acad.Sci.USA / Year: 2024Title: TIFAB regulates the TIFA-TRAF6 signaling pathway involved in innate immunity by forming a heterodimer complex with TIFA. Authors: Nakamura, T. / Ohyama, C. / Sakamoto, M. / Toma, T. / Tateishi, H. / Matsuo, M. / Chirifu, M. / Ikemizu, S. / Morioka, H. / Fujita, M. / Inoue, J.I. / Yamagata, Y. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8wwy.cif.gz 8wwy.cif.gz | 299.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8wwy.ent.gz pdb8wwy.ent.gz | 202.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8wwy.json.gz 8wwy.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ww/8wwy https://data.pdbj.org/pub/pdb/validation_reports/ww/8wwy ftp://data.pdbj.org/pub/pdb/validation_reports/ww/8wwy ftp://data.pdbj.org/pub/pdb/validation_reports/ww/8wwy | HTTPS FTP |
|---|
-Related structure data
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||
| 2 | 
| ||||||||||||
| Unit cell |
| ||||||||||||
| Components on special symmetry positions |
|
- Components
Components
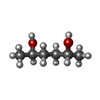
| #1: Protein | Mass: 17577.070 Da / Num. of mol.: 2 / Mutation: C36S Source method: isolated from a genetically manipulated source Source: (gene. exp.)   #2: Protein | Mass: 16167.834 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   #3: Chemical | ChemComp-HX2 / ( #4: Water | ChemComp-HOH / | Has ligand of interest | N | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.51 Å3/Da / Density % sol: 50.9 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop / Details: HEPES, PEG 2000, Hexanediol |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Photon Factory Photon Factory  / Beamline: BL-5A / Wavelength: 1 Å / Beamline: BL-5A / Wavelength: 1 Å |
| Detector | Type: DECTRIS PILATUS3 6M / Detector: PIXEL / Date: Dec 15, 2020 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 1.79→47.04 Å / Num. obs: 65696 / % possible obs: 99.7 % / Redundancy: 19.5 % / Biso Wilson estimate: 31.4 Å2 / CC1/2: 1 / Net I/σ(I): 21.4 |
| Reflection shell | Resolution: 1.79→1.9 Å / Num. unique obs: 10329 / CC1/2: 0.738 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT / Resolution: 1.79→47.04 Å / SU ML: 0.2006 / Cross valid method: FREE R-VALUE / σ(F): 1.34 / Phase error: 22.3703 / Stereochemistry target values: GeoStd + Monomer Library MOLECULAR REPLACEMENT / Resolution: 1.79→47.04 Å / SU ML: 0.2006 / Cross valid method: FREE R-VALUE / σ(F): 1.34 / Phase error: 22.3703 / Stereochemistry target values: GeoStd + Monomer Library
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 40.83 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.79→47.04 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller



 PDBj
PDBj