[English] 日本語
 Yorodumi
Yorodumi- PDB-8v1f: TMPRSS2 complexed with the noncovalent inhibitor 6-amidino-2-napthol -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8v1f | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | TMPRSS2 complexed with the noncovalent inhibitor 6-amidino-2-napthol | |||||||||
 Components Components | (Transmembrane protease serine ...) x 2 | |||||||||
 Keywords Keywords | HYDROLASE/HYDROLASE INHIBITOR / Inhibitor complex / protease / viral entry / Structural Genomics / Structural Genomics Consortium / SGC / HYDROLASE / HYDROLASE-HYDROLASE INHIBITOR complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationtransmembrane protease serine 2 / protein autoprocessing / viral translation / Attachment and Entry / serine-type peptidase activity / Induction of Cell-Cell Fusion / Attachment and Entry / entry receptor-mediated virion attachment to host cell / positive regulation of viral entry into host cell / serine-type endopeptidase activity ...transmembrane protease serine 2 / protein autoprocessing / viral translation / Attachment and Entry / serine-type peptidase activity / Induction of Cell-Cell Fusion / Attachment and Entry / entry receptor-mediated virion attachment to host cell / positive regulation of viral entry into host cell / serine-type endopeptidase activity / proteolysis / extracellular exosome / extracellular region / nucleoplasm / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.19 Å MOLECULAR REPLACEMENT / Resolution: 2.19 Å | |||||||||
 Authors Authors | Fraser, B.J. / Dong, A. / Kutera, M. / Seitova, A. / Li, Y. / Hutchinson, A. / Edwards, A. / Benard, F. / Halabelian, L. / Arrowsmith, C. / Structural Genomics Consortium (SGC) | |||||||||
| Funding support |  Canada, Canada,  United States, 2items United States, 2items
| |||||||||
 Citation Citation |  Journal: J.Med.Chem. / Year: 2025 Journal: J.Med.Chem. / Year: 2025Title: Large Library Docking and Biophysical Analysis of Small-Molecule TMPRSS2 Inhibitors. Authors: Fraser, B.J. / Young, N.J. / Bender, B.J. / Gahbauer, S. / Ilyassov, O. / Wilson, R.P. / Li, Y. / Seitova, A. / Lourenco, A.L. / Chung, D.H. / Bardine, C. / Benard, F. / Shoichet, B.K. / ...Authors: Fraser, B.J. / Young, N.J. / Bender, B.J. / Gahbauer, S. / Ilyassov, O. / Wilson, R.P. / Li, Y. / Seitova, A. / Lourenco, A.L. / Chung, D.H. / Bardine, C. / Benard, F. / Shoichet, B.K. / Craik, C.S. / Arrowsmith, C.H. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8v1f.cif.gz 8v1f.cif.gz | 167.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8v1f.ent.gz pdb8v1f.ent.gz | 127.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8v1f.json.gz 8v1f.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/v1/8v1f https://data.pdbj.org/pub/pdb/validation_reports/v1/8v1f ftp://data.pdbj.org/pub/pdb/validation_reports/v1/8v1f ftp://data.pdbj.org/pub/pdb/validation_reports/v1/8v1f | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  8v04C  9e83C C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
| ||||||||
| Components on special symmetry positions |
|
- Components
Components
-Transmembrane protease serine ... , 2 types, 4 molecules ACBD
| #1: Protein | Mass: 12264.598 Da / Num. of mol.: 2 / Fragment: SRCR domain non-catalytic chain residues 148-254 / Mutation: S251D, R252D, Q253D, S254D, R255K Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: TMPRSS2, PRSS10 / Plasmid: pFHSMP-LIC-C Homo sapiens (human) / Gene: TMPRSS2, PRSS10 / Plasmid: pFHSMP-LIC-CDetails (production host): baculovirus donor vector for secreted protein production Production host:  #2: Protein | Mass: 27856.742 Da / Num. of mol.: 2 / Fragment: Peptidase S1 domain residues 256-492 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: TMPRSS2 / Plasmid: pFHMSP-LIC-C Homo sapiens (human) / Gene: TMPRSS2 / Plasmid: pFHMSP-LIC-CDetails (production host): baculovirus donor vector for secreted protein production Production host:  |
|---|
-Sugars , 2 types, 2 molecules
| #3: Polysaccharide | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose Source method: isolated from a genetically manipulated source |
|---|---|
| #4: Polysaccharide | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta- ...2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose Source method: isolated from a genetically manipulated source |
-Non-polymers , 8 types, 344 molecules 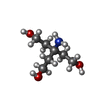


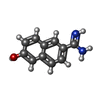











| #5: Chemical | ChemComp-TAM / #6: Chemical | #7: Chemical | ChemComp-EDO / #8: Chemical | #9: Chemical | ChemComp-PEG / | #10: Chemical | #11: Chemical | #12: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.68 Å3/Da / Density % sol: 54.09 % / Description: 2D diamond shaped plates |
|---|---|
| Crystal grow | Temperature: 291 K / Method: vapor diffusion, hanging drop Details: precipitant containing 20% PEG3350, 0.2 M dibasic ammonium citrate. Protein mixed 1uL:1uL protein:precipitant and set as 2 uL hanging drop on glass slides PH range: 4.8 to 5.3 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 24-ID-C / Wavelength: 0.97918 Å / Beamline: 24-ID-C / Wavelength: 0.97918 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: DECTRIS EIGER X 16M / Detector: PIXEL / Date: Jun 11, 2022 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Monochromator: Zr filter / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 0.97918 Å / Relative weight: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 2.19→40 Å / Num. obs: 44382 / % possible obs: 99.7 % / Redundancy: 9 % / CC1/2: 0.984 / CC star: 0.996 / Rmerge(I) obs: 0.226 / Rpim(I) all: 0.076 / Rrim(I) all: 0.239 / Χ2: 1.057 / Net I/σ(I): 5.4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1
|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT / Resolution: 2.19→38.67 Å / Cor.coef. Fo:Fc: 0.966 / Cor.coef. Fo:Fc free: 0.945 / SU B: 7.001 / SU ML: 0.166 / Cross valid method: THROUGHOUT / ESU R: 0.204 / ESU R Free: 0.18 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS MOLECULAR REPLACEMENT / Resolution: 2.19→38.67 Å / Cor.coef. Fo:Fc: 0.966 / Cor.coef. Fo:Fc free: 0.945 / SU B: 7.001 / SU ML: 0.166 / Cross valid method: THROUGHOUT / ESU R: 0.204 / ESU R Free: 0.18 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 40.183 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: 1 / Resolution: 2.19→38.67 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller


 PDBj
PDBj








