[English] 日本語
 Yorodumi
Yorodumi- PDB-8thl: Cryo-EM structure of epinephrine-bound alpha-1A-adrenergic recept... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8thl | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of epinephrine-bound alpha-1A-adrenergic receptor in complex with heterotrimeric Gq-protein | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | SIGNALING PROTEIN / Alpha-1A-adrenergic receptor / Epinephrine | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of heart rate involved in baroreceptor response to increased systemic arterial blood pressure / alpha1-adrenergic receptor activity / norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure / positive regulation of heart rate by epinephrine-norepinephrine / positive regulation of the force of heart contraction by epinephrine-norepinephrine / pilomotor reflex / phospholipase C-activating adrenergic receptor signaling pathway / neuron-glial cell signaling / cell growth involved in cardiac muscle cell development / negative regulation of adenylate cyclase-activating adrenergic receptor signaling pathway ...negative regulation of heart rate involved in baroreceptor response to increased systemic arterial blood pressure / alpha1-adrenergic receptor activity / norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure / positive regulation of heart rate by epinephrine-norepinephrine / positive regulation of the force of heart contraction by epinephrine-norepinephrine / pilomotor reflex / phospholipase C-activating adrenergic receptor signaling pathway / neuron-glial cell signaling / cell growth involved in cardiac muscle cell development / negative regulation of adenylate cyclase-activating adrenergic receptor signaling pathway / Fatty Acids bound to GPR40 (FFAR1) regulate insulin secretion / Acetylcholine regulates insulin secretion / phospholipase C-activating G protein-coupled glutamate receptor signaling pathway / positive regulation of action potential / positive regulation of smooth muscle contraction / PLC beta mediated events / phospholipase C-activating serotonin receptor signaling pathway / negative regulation of calcium ion-dependent exocytosis / G protein-coupled adenosine receptor signaling pathway / entrainment of circadian clock / regulation of platelet activation / negative regulation of adenylate cyclase activity / positive regulation of urine volume / adult heart development / positive regulation of neural precursor cell proliferation / negative regulation of synaptic transmission / regulation of canonical Wnt signaling pathway / Adrenoceptors / glutamate receptor signaling pathway / gamma-aminobutyric acid signaling pathway / positive regulation of cardiac muscle hypertrophy / neuronal dense core vesicle / regulation of calcium ion transport / negative regulation of apoptotic signaling pathway / phototransduction, visible light / smooth muscle contraction / PKA activation in glucagon signalling / developmental growth / hair follicle placode formation / photoreceptor outer segment / neuropeptide signaling pathway / postsynaptic cytosol / D1 dopamine receptor binding / intracellular transport / vascular endothelial cell response to laminar fluid shear stress / positive regulation of vasoconstriction / renal water homeostasis / viral release from host cell by cytolysis / activation of adenylate cyclase activity / Hedgehog 'off' state / Adenylate cyclase inhibitory pathway / positive regulation of vascular associated smooth muscle cell proliferation / response to hormone / negative regulation of protein kinase activity / adenylate cyclase-activating adrenergic receptor signaling pathway / response to prostaglandin E / enzyme regulator activity / peptidoglycan catabolic process / response to nutrient / positive regulation of cardiac muscle contraction / negative regulation of autophagy / cellular response to glucagon stimulus / regulation of insulin secretion / GTPase activator activity / hippocampal mossy fiber to CA3 synapse / adenylate cyclase activator activity / positive regulation of superoxide anion generation / Turbulent (oscillatory, disturbed) flow shear stress activates signaling by PIEZO1 and integrins in endothelial cells / trans-Golgi network membrane / positive regulation of synaptic transmission, GABAergic / Regulation of insulin secretion / negative regulation of inflammatory response to antigenic stimulus / G protein-coupled receptor binding / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / bone development / caveola / G-protein beta/gamma-subunit complex binding / platelet aggregation / Olfactory Signaling Pathway / Activation of the phototransduction cascade / cognition / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / G-protein activation / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Prostacyclin signalling through prostacyclin receptor / G beta:gamma signalling through CDC42 / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / Sensory perception of sweet, bitter, and umami (glutamate) taste / photoreceptor disc membrane / Glucagon-type ligand receptors / cell wall macromolecule catabolic process / blood coagulation Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) Enterobacteria phage T4 (virus) Enterobacteria phage T4 (virus) | |||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Su, M. / Huang, X.Y. | |||||||||
| Funding support |  United States, 2items United States, 2items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural basis of agonist specificity of α-adrenergic receptor. Authors: Minfei Su / Jinan Wang / Guoqing Xiang / Hung Nguyen Do / Joshua Levitz / Yinglong Miao / Xin-Yun Huang /  Abstract: α-adrenergic receptors (α-ARs) play critical roles in the cardiovascular and nervous systems where they regulate blood pressure, cognition, and metabolism. However, the lack of specific agonists ...α-adrenergic receptors (α-ARs) play critical roles in the cardiovascular and nervous systems where they regulate blood pressure, cognition, and metabolism. However, the lack of specific agonists for all α subtypes has limited our understanding of the physiological roles of different α-AR subtypes, and led to the stagnancy in agonist-based drug development for these receptors. Here we report cryo-EM structures of α-AR in complex with heterotrimeric G-proteins and either the endogenous common agonist epinephrine or the α-AR-specific synthetic agonist A61603. These structures provide molecular insights into the mechanisms underlying the discrimination between α-AR and α-AR by A61603. Guided by the structures and corresponding molecular dynamics simulations, we engineer α-AR mutants that are not responsive to A61603, and α-AR mutants that can be potently activated by A61603. Together, these findings advance our understanding of the agonist specificity for α-ARs at the molecular level, opening the possibility of rational design of subtype-specific agonists. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8thl.cif.gz 8thl.cif.gz | 223.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8thl.ent.gz pdb8thl.ent.gz | 165.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8thl.json.gz 8thl.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  8thl_validation.pdf.gz 8thl_validation.pdf.gz | 1.2 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  8thl_full_validation.pdf.gz 8thl_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  8thl_validation.xml.gz 8thl_validation.xml.gz | 39 KB | Display | |
| Data in CIF |  8thl_validation.cif.gz 8thl_validation.cif.gz | 57.9 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/th/8thl https://data.pdbj.org/pub/pdb/validation_reports/th/8thl ftp://data.pdbj.org/pub/pdb/validation_reports/th/8thl ftp://data.pdbj.org/pub/pdb/validation_reports/th/8thl | HTTPS FTP |
-Related structure data
| Related structure data |  41268MC  8thkC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Guanine nucleotide-binding protein ... , 3 types, 3 molecules ABG
| #1: Protein | Mass: 28084.832 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: GNAI2, GNAI2B, GNAS, GNAS1, GSP, GNAQ, GAQ / Production host: Homo sapiens (human) / Gene: GNAI2, GNAI2B, GNAS, GNAS1, GSP, GNAQ, GAQ / Production host:  References: UniProt: P04899, UniProt: P63092, UniProt: P50148 |
|---|---|
| #2: Protein | Mass: 39418.086 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: GNB1 / Production host: Homo sapiens (human) / Gene: GNB1 / Production host:  |
| #3: Protein | Mass: 7861.143 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: GNG2 / Production host: Homo sapiens (human) / Gene: GNG2 / Production host:  |
-Antibody / Protein / Non-polymers , 3 types, 3 molecules FR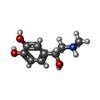

| #4: Antibody | Mass: 28668.922 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Production host: Homo sapiens (human) / Production host:  |
|---|---|
| #5: Protein | Mass: 56485.246 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Enterobacteria phage T4 (virus), (gene. exp.) Enterobacteria phage T4 (virus), (gene. exp.)  Homo sapiens (human) Homo sapiens (human)Gene: E, ADRA1A, ADRA1C / Production host:  |
| #6: Chemical | ChemComp-ALE / |
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Epinephrine-bound alpha-1A-adrenergic receptor in complex with heterotrimeric Gq-protein Type: COMPLEX / Entity ID: #1-#5 / Source: RECOMBINANT |
|---|---|
| Molecular weight | Value: 0.16 MDa / Experimental value: NO |
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Source (recombinant) | Organism:  |
| Buffer solution | pH: 7 |
| Specimen | Conc.: 2.5 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal defocus max: 1800 nm / Nominal defocus min: 1000 nm |
| Image recording | Electron dose: 52 e/Å2 / Film or detector model: GATAN K3 (6k x 4k) |
- Processing
Processing
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3D reconstruction | Resolution: 3.1 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 219834 / Symmetry type: POINT | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller



 PDBj
PDBj































