[English] 日本語
 Yorodumi
Yorodumi- EMDB-41267: Cryo-EM structure of A61603-bound alpha-1A-adrenergic receptor in... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
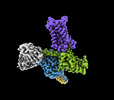
| Title | Cryo-EM structure of A61603-bound alpha-1A-adrenergic receptor in complex with heterotrimeric Gq-protein | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Alpha-1A-adrenergic receptor / A61603 / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of heart rate involved in baroreceptor response to increased systemic arterial blood pressure / alpha1-adrenergic receptor activity / norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure / positive regulation of heart rate by epinephrine-norepinephrine / positive regulation of the force of heart contraction by epinephrine-norepinephrine / pilomotor reflex / phospholipase C-activating adrenergic receptor signaling pathway / neuron-glial cell signaling / cell growth involved in cardiac muscle cell development / Fatty Acids bound to GPR40 (FFAR1) regulate insulin secretion ...negative regulation of heart rate involved in baroreceptor response to increased systemic arterial blood pressure / alpha1-adrenergic receptor activity / norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure / positive regulation of heart rate by epinephrine-norepinephrine / positive regulation of the force of heart contraction by epinephrine-norepinephrine / pilomotor reflex / phospholipase C-activating adrenergic receptor signaling pathway / neuron-glial cell signaling / cell growth involved in cardiac muscle cell development / Fatty Acids bound to GPR40 (FFAR1) regulate insulin secretion / negative regulation of adenylate cyclase-activating adrenergic receptor signaling pathway / Acetylcholine regulates insulin secretion / phospholipase C-activating G protein-coupled glutamate receptor signaling pathway / positive regulation of action potential / positive regulation of smooth muscle contraction / PLC beta mediated events / phospholipase C-activating serotonin receptor signaling pathway / negative regulation of calcium ion-dependent exocytosis / regulation of platelet activation / entrainment of circadian clock / G protein-coupled adenosine receptor signaling pathway / negative regulation of adenylate cyclase activity / positive regulation of urine volume / adult heart development / positive regulation of neural precursor cell proliferation / negative regulation of synaptic transmission / Adrenoceptors / regulation of canonical Wnt signaling pathway / glutamate receptor signaling pathway / positive regulation of cardiac muscle hypertrophy / gamma-aminobutyric acid signaling pathway / positive regulation of vasoconstriction / regulation of calcium ion transport / smooth muscle contraction / phototransduction, visible light / negative regulation of apoptotic signaling pathway / PKA activation in glucagon signalling / developmental growth / hair follicle placode formation / photoreceptor outer segment / D1 dopamine receptor binding / intracellular transport / neuropeptide signaling pathway / postsynaptic cytosol / neuronal dense core vesicle / vascular endothelial cell response to laminar fluid shear stress / viral release from host cell by cytolysis / renal water homeostasis / activation of adenylate cyclase activity / Hedgehog 'off' state / positive regulation of vascular associated smooth muscle cell proliferation / positive regulation of superoxide anion generation / adenylate cyclase-activating adrenergic receptor signaling pathway / response to hormone / enzyme regulator activity / Adenylate cyclase inhibitory pathway / response to prostaglandin E / peptidoglycan catabolic process / positive regulation of cardiac muscle contraction / response to nutrient / regulation of insulin secretion / cellular response to glucagon stimulus / negative regulation of autophagy / GTPase activator activity / hippocampal mossy fiber to CA3 synapse / Turbulent (oscillatory, disturbed) flow shear stress activates signaling by PIEZO1 and integrins in endothelial cells / adenylate cyclase activator activity / trans-Golgi network membrane / positive regulation of synaptic transmission, GABAergic / Regulation of insulin secretion / negative regulation of inflammatory response to antigenic stimulus / G protein-coupled receptor binding / bone development / caveola / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / platelet aggregation / cognition / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Activation of the phototransduction cascade / cell wall macromolecule catabolic process / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / blood coagulation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / lysozyme / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.6 Å | |||||||||
 Authors Authors | Su M / Huang XY | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural basis of agonist specificity of α-adrenergic receptor. Authors: Minfei Su / Jinan Wang / Guoqing Xiang / Hung Nguyen Do / Joshua Levitz / Yinglong Miao / Xin-Yun Huang /  Abstract: α-adrenergic receptors (α-ARs) play critical roles in the cardiovascular and nervous systems where they regulate blood pressure, cognition, and metabolism. However, the lack of specific agonists ...α-adrenergic receptors (α-ARs) play critical roles in the cardiovascular and nervous systems where they regulate blood pressure, cognition, and metabolism. However, the lack of specific agonists for all α subtypes has limited our understanding of the physiological roles of different α-AR subtypes, and led to the stagnancy in agonist-based drug development for these receptors. Here we report cryo-EM structures of α-AR in complex with heterotrimeric G-proteins and either the endogenous common agonist epinephrine or the α-AR-specific synthetic agonist A61603. These structures provide molecular insights into the mechanisms underlying the discrimination between α-AR and α-AR by A61603. Guided by the structures and corresponding molecular dynamics simulations, we engineer α-AR mutants that are not responsive to A61603, and α-AR mutants that can be potently activated by A61603. Together, these findings advance our understanding of the agonist specificity for α-ARs at the molecular level, opening the possibility of rational design of subtype-specific agonists. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41267.map.gz emd_41267.map.gz | 64.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41267-v30.xml emd-41267-v30.xml emd-41267.xml emd-41267.xml | 22 KB 22 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_41267.png emd_41267.png | 64 KB | ||
| Filedesc metadata |  emd-41267.cif.gz emd-41267.cif.gz | 6.8 KB | ||
| Others |  emd_41267_additional_1.map.gz emd_41267_additional_1.map.gz emd_41267_half_map_1.map.gz emd_41267_half_map_1.map.gz emd_41267_half_map_2.map.gz emd_41267_half_map_2.map.gz | 33.2 MB 59.4 MB 59.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41267 http://ftp.pdbj.org/pub/emdb/structures/EMD-41267 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41267 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41267 | HTTPS FTP |
-Validation report
| Summary document |  emd_41267_validation.pdf.gz emd_41267_validation.pdf.gz | 765.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_41267_full_validation.pdf.gz emd_41267_full_validation.pdf.gz | 764.9 KB | Display | |
| Data in XML |  emd_41267_validation.xml.gz emd_41267_validation.xml.gz | 13.3 KB | Display | |
| Data in CIF |  emd_41267_validation.cif.gz emd_41267_validation.cif.gz | 15.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41267 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41267 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41267 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41267 | HTTPS FTP |
-Related structure data
| Related structure data |  8thkMC  8thlC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41267.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41267.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.856 Å | ||||||||||||||||||||||||||||||||||||
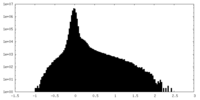
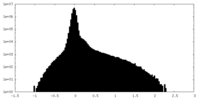
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Receptor and Ga focused map
| File | emd_41267_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Receptor and Ga focused map | ||||||||||||
| Projections & Slices |
| ||||||||||||
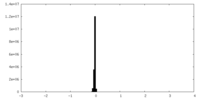
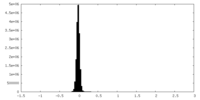
| Density Histograms |
-Half map: #2
| File | emd_41267_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
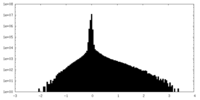
| Density Histograms |
-Half map: #1
| File | emd_41267_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
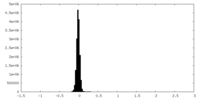
| Density Histograms |
- Sample components
Sample components
-Entire : A61603-bound alpha-1A-adrenergic receptor in complex with heterot...
| Entire | Name: A61603-bound alpha-1A-adrenergic receptor in complex with heterotrimeric Gq-protein |
|---|---|
| Components |
|
-Supramolecule #1: A61603-bound alpha-1A-adrenergic receptor in complex with heterot...
| Supramolecule | Name: A61603-bound alpha-1A-adrenergic receptor in complex with heterotrimeric Gq-protein type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 160 KDa |
-Macromolecule #1: Guanine nucleotide-binding protein G(i) subunit alpha-2,Guanine n...
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-2,Guanine nucleotide-binding protein G(s) subunit alpha isoforms short,Guanine nucleotide-binding protein G(q) subunit alpha type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 28.084832 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSTVSAEDK AAAERSKMID KNLREDGEKA RRTLRLLLLG ADNSGKSTIV KQMRILHGGS GGSGGTSGIF ETKFQVDKVN FHMFDVGGQ RDERRKWIQC FNDVTAIIFV VDSSDYNRLQ EALNDFKSIW NNRWLRTISV ILFLNKQDLL AEKVLAGKSK I EDYFPEFA ...String: MGSTVSAEDK AAAERSKMID KNLREDGEKA RRTLRLLLLG ADNSGKSTIV KQMRILHGGS GGSGGTSGIF ETKFQVDKVN FHMFDVGGQ RDERRKWIQC FNDVTAIIFV VDSSDYNRLQ EALNDFKSIW NNRWLRTISV ILFLNKQDLL AEKVLAGKSK I EDYFPEFA RYTTPEDATP EPGEDPRVTR AKYFIRKEFV DISTASGDGR HICYPHFTCA VDTENARRIF NDCKDIILQM NL REYNLV UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-2, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, Guanine nucleotide-binding protein G(q) subunit alpha |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.418086 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHLEV LFQGPGSSGS ELDQLRQEAE QLKNQIRDAR KACADATLSQ ITNNIDPVGR IQMRTRRTLR GHLAKIYAMH WGTDSRLLV SASQDGKLII WDSYTTNKVH AIPLRSSWVM TCAYAPSGNY VACGGLDNIC SIYNLKTREG NVRVSRELAG H TGYLSCCR ...String: MHHHHHHLEV LFQGPGSSGS ELDQLRQEAE QLKNQIRDAR KACADATLSQ ITNNIDPVGR IQMRTRRTLR GHLAKIYAMH WGTDSRLLV SASQDGKLII WDSYTTNKVH AIPLRSSWVM TCAYAPSGNY VACGGLDNIC SIYNLKTREG NVRVSRELAG H TGYLSCCR FLDDNQIVTS SGDTTCALWD IETGQQTTTF TGHTGDVMSL SLAPDTRLFV SGACDASAKL WDVREGMCRQ TF TGHESDI NAICFFPNGN AFATGSDDAT CRLFDLRADQ ELMTYSHDNI ICGITSVSFS KSGRLLLAGY DDFNCNVWDA LKA DRAGVL AGHDNRVSCL GVTDDGMAVA TGSWDSFLKI WN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: Single fab chain (scFv16)
| Macromolecule | Name: Single fab chain (scFv16) / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 28.668922 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL KAAALEVLFQ GPHHHHHHHH |
-Macromolecule #5: Endolysin,Alpha-1A adrenergic receptor
| Macromolecule | Name: Endolysin,Alpha-1A adrenergic receptor / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO / EC number: lysozyme |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 56.485246 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDKNIFEML RIDEGLRLKI YKDTEGYYTI GIGHLLTKSP SLNAAKSELD KAIGRNTNGV ITKDEAEKL FNQDVDAAVR GILRNAKLKP VYDSLDAVRR AALINMVFQM GETGVAGFTN SLRMLQQKRW DEAAVNLAKS R WYNQTPNR ...String: MKTIIALSYI FCLVFADYKD DDDKNIFEML RIDEGLRLKI YKDTEGYYTI GIGHLLTKSP SLNAAKSELD KAIGRNTNGV ITKDEAEKL FNQDVDAAVR GILRNAKLKP VYDSLDAVRR AALINMVFQM GETGVAGFTN SLRMLQQKRW DEAAVNLAKS R WYNQTPNR AKRVITTFRT GTWDAYAAAT QPPAPVNISK AILLGVILGG LILFGVLGNI LVILSVACHR HLHSVTHYYI VN LAVADLL LTSTVLPFSA IFEVLGYWAF GRVFCNIWAA VDVLCCTASI MGLCIISIDR YIGVSYPLRY PTIVTQRRGL MAL LCVWAL SLVISIGPLF GWRQPAPEDE TICQINEEPG YVLFSALGSF YLPLAIILVM YCRVYVVAKR ESRGLKSGLK TDKS HFSVR LLKFSREKKA AKTLGIVVGC FVLCWLPFFL VMPIGSFFPD FKPSETVFKI VFWLGYLNSC INPIIYPCSS QEFKK AFQN VLRIQCLCRK QASLEVLFQ UniProtKB: Endolysin, Alpha-1A adrenergic receptor |
-Macromolecule #6: N-[(5S)-5-(4,5-dihydro-1H-imidazol-2-yl)-2-hydroxy-5,6,7,8-tetrah...
| Macromolecule | Name: N-[(5S)-5-(4,5-dihydro-1H-imidazol-2-yl)-2-hydroxy-5,6,7,8-tetrahydronaphthalen-1-yl]methanesulfonamide type: ligand / ID: 6 / Number of copies: 1 / Formula: CGZ |
|---|---|
| Molecular weight | Theoretical: 309.384 Da |
| Chemical component information | 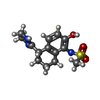 ChemComp-CGZ: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL |
|---|---|
| Buffer | pH: 7 |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.6 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 360489 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller
































 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)