[English] 日本語
 Yorodumi
Yorodumi- PDB-8p34: Tau filaments extracted from human brain with the DeltaK281 mutat... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8p34 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Tau filaments extracted from human brain with the DeltaK281 mutation in MAPT | ||||||
 Components Components | Microtubule-associated protein tau | ||||||
 Keywords Keywords | PROTEIN FIBRIL / DeltaK281 / Tau / Amyloid filament / Cross-beta-sheet | ||||||
| Function / homology |  Function and homology information Function and homology informationplus-end-directed organelle transport along microtubule / histone-dependent DNA binding / negative regulation of protein localization to mitochondrion / neurofibrillary tangle / microtubule lateral binding / axonal transport / tubulin complex / positive regulation of protein localization to synapse / phosphatidylinositol bisphosphate binding / generation of neurons ...plus-end-directed organelle transport along microtubule / histone-dependent DNA binding / negative regulation of protein localization to mitochondrion / neurofibrillary tangle / microtubule lateral binding / axonal transport / tubulin complex / positive regulation of protein localization to synapse / phosphatidylinositol bisphosphate binding / generation of neurons / rRNA metabolic process / axonal transport of mitochondrion / regulation of mitochondrial fission / axon development / regulation of microtubule-based movement / regulation of chromosome organization / central nervous system neuron development / intracellular distribution of mitochondria / minor groove of adenine-thymine-rich DNA binding / lipoprotein particle binding / microtubule polymerization / negative regulation of mitochondrial membrane potential / regulation of microtubule polymerization / dynactin binding / apolipoprotein binding / main axon / protein polymerization / axolemma / Caspase-mediated cleavage of cytoskeletal proteins / regulation of microtubule polymerization or depolymerization / negative regulation of mitochondrial fission / glial cell projection / neurofibrillary tangle assembly / positive regulation of axon extension / regulation of cellular response to heat / Activation of AMPK downstream of NMDARs / positive regulation of superoxide anion generation / positive regulation of protein localization / cellular response to brain-derived neurotrophic factor stimulus / supramolecular fiber organization / regulation of long-term synaptic depression / positive regulation of microtubule polymerization / cytoplasmic microtubule organization / synapse assembly / regulation of calcium-mediated signaling / somatodendritic compartment / axon cytoplasm / astrocyte activation / phosphatidylinositol binding / nuclear periphery / enzyme inhibitor activity / stress granule assembly / protein phosphatase 2A binding / regulation of microtubule cytoskeleton organization / cellular response to reactive oxygen species / microglial cell activation / Hsp90 protein binding / cellular response to nerve growth factor stimulus / protein homooligomerization / synapse organization / PKR-mediated signaling / regulation of synaptic plasticity / regulation of autophagy / SH3 domain binding / response to lead ion / microtubule cytoskeleton organization / memory / neuron projection development / cytoplasmic ribonucleoprotein granule / cell-cell signaling / single-stranded DNA binding / protein-folding chaperone binding / cellular response to heat / microtubule cytoskeleton / actin binding / growth cone / cell body / double-stranded DNA binding / microtubule binding / protein-macromolecule adaptor activity / sequence-specific DNA binding / amyloid fibril formation / dendritic spine / microtubule / learning or memory / neuron projection / membrane raft / negative regulation of gene expression / axon / neuronal cell body / DNA damage response / dendrite / protein kinase binding / enzyme binding / mitochondrion / DNA binding / RNA binding / extracellular region / identical protein binding / nucleus Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method | ELECTRON MICROSCOPY / helical reconstruction / cryo EM / Resolution: 2.61 Å | ||||||
 Authors Authors | Schweighauser, M. / Garringer, H.J. / Klingstedt, T. / Masuda-Suzukake, M. / Murrell, J.R. / Vidal, R. / Scheres, S.H.W. / Goedert, M. / Ghetti, B. / Newell, K.L. | ||||||
| Funding support |  United Kingdom, 1items United Kingdom, 1items
| ||||||
 Citation Citation |  Journal: Acta Neuropathol / Year: 2023 Journal: Acta Neuropathol / Year: 2023Title: Mutation ∆K281 in MAPT causes Pick's disease. Authors: Manuel Schweighauser / Holly J Garringer / Therése Klingstedt / K Peter R Nilsson / Masami Masuda-Suzukake / Jill R Murrell / Shannon L Risacher / Ruben Vidal / Sjors H W Scheres / Michel ...Authors: Manuel Schweighauser / Holly J Garringer / Therése Klingstedt / K Peter R Nilsson / Masami Masuda-Suzukake / Jill R Murrell / Shannon L Risacher / Ruben Vidal / Sjors H W Scheres / Michel Goedert / Bernardino Ghetti / Kathy L Newell /     Abstract: Two siblings with deletion mutation ∆K281 in MAPT developed frontotemporal dementia. At autopsy, numerous inclusions of hyperphosphorylated 3R Tau were present in neurons and glial cells of ...Two siblings with deletion mutation ∆K281 in MAPT developed frontotemporal dementia. At autopsy, numerous inclusions of hyperphosphorylated 3R Tau were present in neurons and glial cells of neocortex and some subcortical regions, including hippocampus, caudate/putamen and globus pallidus. The inclusions were argyrophilic with Bodian silver, but not with Gallyas-Braak silver. They were not labelled by an antibody specific for tau phosphorylated at S262 and/or S356. The inclusions were stained by luminescent conjugated oligothiophene HS-84, but not by bTVBT4. Electron cryo-microscopy revealed that the core of tau filaments was made of residues K254-F378 of 3R Tau and was indistinguishable from that of Pick's disease. We conclude that MAPT mutation ∆K281 causes Pick's disease. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8p34.cif.gz 8p34.cif.gz | 30.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8p34.ent.gz pdb8p34.ent.gz | 17.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8p34.json.gz 8p34.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/p3/8p34 https://data.pdbj.org/pub/pdb/validation_reports/p3/8p34 ftp://data.pdbj.org/pub/pdb/validation_reports/p3/8p34 ftp://data.pdbj.org/pub/pdb/validation_reports/p3/8p34 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  17383MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 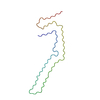
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein | Mass: 10135.665 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) / References: UniProt: P10636 Homo sapiens (human) / References: UniProt: P10636 |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: FILAMENT / 3D reconstruction method: helical reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Tau filaments extracted from human brain with DeltaK281 mutation Type: TISSUE / Entity ID: all / Source: NATURAL |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Organ: Brain / Tissue: Frontal cortex grey matter Homo sapiens (human) / Organ: Brain / Tissue: Frontal cortex grey matter |
| Buffer solution | pH: 7.4 / Details: 20 mM Tris-HCL, 100 mM NaCl |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Specimen support | Grid material: GOLD / Grid mesh size: 300 divisions/in. / Grid type: Quantifoil R1.2/1.3 |
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 277.15 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal defocus max: 2800 nm / Nominal defocus min: 1800 nm / Alignment procedure: COMA FREE |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Average exposure time: 1 sec. / Electron dose: 37.7 e/Å2 / Film or detector model: GATAN K3 (6k x 4k) / Num. of real images: 41385 |
| EM imaging optics | Phase plate: OTHER |
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: NONE | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Helical symmerty | Angular rotation/subunit: -0.73 ° / Axial rise/subunit: 4.89 Å / Axial symmetry: C1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 1000880 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 2.61 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 258445 / Symmetry type: HELICAL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Atomic model building | PDB-ID: 6GX5 Accession code: 6GX5 / Source name: PDB / Type: experimental model | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | Resolution: 2.61→124.62 Å / Cor.coef. Fo:Fc: 0.847 / SU B: 5.769 / SU ML: 0.119 / ESU R: 0.106 Stereochemistry target values: MAXIMUM LIKELIHOOD WITH PHASES Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: PARAMETERS FOR MASK CACLULATION | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 53.833 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: 1 / Total: 711 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller


 PDBj
PDBj