+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: PDB / ID: 8fno | ||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Structure of E138K/G140A/Q148R HIV-1 intasome with Dolutegravir bound | ||||||||||||||||||||||||||||||||||||||||||||||||
 要素 要素 |
| ||||||||||||||||||||||||||||||||||||||||||||||||
 キーワード キーワード | VIRAL PROTEIN/DNA/INHIBITOR / Integrase / Nucleoprotein complex / Inhibitor / Drug resistance / VIRAL PROTEIN-DNA-INHIBITOR complex | ||||||||||||||||||||||||||||||||||||||||||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報Depolymerization of the Nuclear Lamina / Nuclear Envelope Breakdown / lamin binding / Initiation of Nuclear Envelope (NE) Reformation / RHOD GTPase cycle / RHOF GTPase cycle / nuclear inner membrane / RHOC GTPase cycle / RHOJ GTPase cycle / CDC42 GTPase cycle ...Depolymerization of the Nuclear Lamina / Nuclear Envelope Breakdown / lamin binding / Initiation of Nuclear Envelope (NE) Reformation / RHOD GTPase cycle / RHOF GTPase cycle / nuclear inner membrane / RHOC GTPase cycle / RHOJ GTPase cycle / CDC42 GTPase cycle / RHOG GTPase cycle / RHOA GTPase cycle / RAC2 GTPase cycle / RAC3 GTPase cycle / RAC1 GTPase cycle / HIV-1 retropepsin / symbiont-mediated activation of host apoptosis / retroviral ribonuclease H / exoribonuclease H / exoribonuclease H activity / host multivesicular body / DNA integration / viral genome integration into host DNA / RNA-directed DNA polymerase / establishment of integrated proviral latency / telomerase activity / viral penetration into host nucleus / RNA stem-loop binding / RNA-DNA hybrid ribonuclease activity / 転移酵素; リンを含む基を移すもの; 核酸を移すもの / nuclear envelope / host cell / viral nucleocapsid / DNA recombination / nuclear membrane / DNA-directed DNA polymerase / aspartic-type endopeptidase activity / 加水分解酵素; エステル加水分解酵素 / DNA-directed DNA polymerase activity / symbiont-mediated suppression of host gene expression / symbiont entry into host cell / viral translational frameshifting / lipid binding / host cell nucleus / host cell plasma membrane / virion membrane / structural molecule activity / proteolysis / DNA binding / zinc ion binding / nucleus / membrane / cytoplasm 類似検索 - 分子機能 | ||||||||||||||||||||||||||||||||||||||||||||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト)  Human immunodeficiency virus 1 (ヒト免疫不全ウイルス) Human immunodeficiency virus 1 (ヒト免疫不全ウイルス) | ||||||||||||||||||||||||||||||||||||||||||||||||
| 手法 | 電子顕微鏡法 / 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 2.46 Å | ||||||||||||||||||||||||||||||||||||||||||||||||
 データ登録者 データ登録者 | Shan, Z.L. / Passos, D.O. / Strutzenberg, T.S. / Li, M. / Lyumkis, D. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 資金援助 |  米国, 5件 米国, 5件
| ||||||||||||||||||||||||||||||||||||||||||||||||
 引用 引用 |  ジャーナル: Sci Adv / 年: 2023 ジャーナル: Sci Adv / 年: 2023タイトル: Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants. 著者: Min Li / Dario Oliveira Passos / Zelin Shan / Steven J Smith / Qinfang Sun / Avik Biswas / Indrani Choudhuri / Timothy S Strutzenberg / Allan Haldane / Nanjie Deng / Zhaoyang Li / Xue Zhi ...著者: Min Li / Dario Oliveira Passos / Zelin Shan / Steven J Smith / Qinfang Sun / Avik Biswas / Indrani Choudhuri / Timothy S Strutzenberg / Allan Haldane / Nanjie Deng / Zhaoyang Li / Xue Zhi Zhao / Lorenzo Briganti / Mamuka Kvaratskhelia / Terrence R Burke / Ronald M Levy / Stephen H Hughes / Robert Craigie / Dmitry Lyumkis /  要旨: HIV-1 infection depends on the integration of viral DNA into host chromatin. Integration is mediated by the viral enzyme integrase and is blocked by integrase strand transfer inhibitors (INSTIs), ...HIV-1 infection depends on the integration of viral DNA into host chromatin. Integration is mediated by the viral enzyme integrase and is blocked by integrase strand transfer inhibitors (INSTIs), first-line antiretroviral therapeutics widely used in the clinic. Resistance to even the best INSTIs is a problem, and the mechanisms of resistance are poorly understood. Here, we analyze combinations of the mutations E138K, G140A/S, and Q148H/K/R, which confer resistance to INSTIs. The investigational drug 4d more effectively inhibited the mutants compared with the approved drug Dolutegravir (DTG). We present 11 new cryo-EM structures of drug-resistant HIV-1 intasomes bound to DTG or 4d, with better than 3-Å resolution. These structures, complemented with free energy simulations, virology, and enzymology, explain the mechanisms of DTG resistance involving E138K + G140A/S + Q148H/K/R and show why 4d maintains potency better than DTG. These data establish a foundation for further development of INSTIs that potently inhibit resistant forms in integrase. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 構造ビューア | 分子:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- ダウンロードとリンク
ダウンロードとリンク
- ダウンロード
ダウンロード
| PDBx/mmCIF形式 |  8fno.cif.gz 8fno.cif.gz | 337.7 KB | 表示 |  PDBx/mmCIF形式 PDBx/mmCIF形式 |
|---|---|---|---|---|
| PDB形式 |  pdb8fno.ent.gz pdb8fno.ent.gz | 253.1 KB | 表示 |  PDB形式 PDB形式 |
| PDBx/mmJSON形式 |  8fno.json.gz 8fno.json.gz | ツリー表示 |  PDBx/mmJSON形式 PDBx/mmJSON形式 | |
| その他 |  その他のダウンロード その他のダウンロード |
-検証レポート
| アーカイブディレクトリ |  https://data.pdbj.org/pub/pdb/validation_reports/fn/8fno https://data.pdbj.org/pub/pdb/validation_reports/fn/8fno ftp://data.pdbj.org/pub/pdb/validation_reports/fn/8fno ftp://data.pdbj.org/pub/pdb/validation_reports/fn/8fno | HTTPS FTP |
|---|
-関連構造データ
| 関連構造データ |  29320MC  8fn7C  8fndC  8fngC  8fnhC  8fnjC  8fnlC  8fnmC  8fnnC  8fnpC  8fnqC C: 同じ文献を引用 ( M: このデータのモデリングに利用したマップデータ |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
- 集合体
集合体
| 登録構造単位 | 
|
|---|---|
| 1 |
|
- 要素
要素
-タンパク質 , 1種, 8分子 ABCDGHIJ
| #1: タンパク質 | 分子量: 39941.512 Da / 分子数: 8 / 変異: E138K,G140A,Q148R / 由来タイプ: 組換発現 由来: (組換発現)  Homo sapiens (ヒト), (組換発現) Homo sapiens (ヒト), (組換発現)   Human immunodeficiency virus 1 (ヒト免疫不全ウイルス) Human immunodeficiency virus 1 (ヒト免疫不全ウイルス)遺伝子: TMPO, LAP2, gag-pol / 発現宿主:  参照: UniProt: P42167, UniProt: P12497, 転移酵素; リンを含む基を移すもの; 核酸を移すもの, 加水分解酵素; エステル加水分解酵素 |
|---|
-DNA鎖 , 2種, 4分子 EKFL
| #2: DNA鎖 | 分子量: 8188.271 Da / 分子数: 2 / 由来タイプ: 合成 由来: (合成)   Human immunodeficiency virus 1 (ヒト免疫不全ウイルス) Human immunodeficiency virus 1 (ヒト免疫不全ウイルス)#3: DNA鎖 | 分子量: 7773.023 Da / 分子数: 2 / 由来タイプ: 合成 由来: (合成)   Human immunodeficiency virus 1 (ヒト免疫不全ウイルス) Human immunodeficiency virus 1 (ヒト免疫不全ウイルス) |
|---|
-非ポリマー , 4種, 898分子 

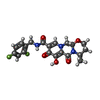




| #4: 化合物 | ChemComp-MG / #5: 化合物 | #6: 化合物 | #7: 水 | ChemComp-HOH / | |
|---|
-詳細
| 研究の焦点であるリガンドがあるか | Y |
|---|---|
| Has protein modification | N |
-実験情報
-実験
| 実験 | 手法: 電子顕微鏡法 |
|---|---|
| EM実験 | 試料の集合状態: PARTICLE / 3次元再構成法: 単粒子再構成法 |
- 試料調製
試料調製
| 構成要素 | 名称: E138K/G140A/Q148R HIV-1 intasome bound to dolutegravir タイプ: COMPLEX / Entity ID: #1-#3 / 由来: MULTIPLE SOURCES | |||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 分子量 | 値: 0.5 MDa / 実験値: NO | |||||||||||||||||||||||||||||||||||||||||||||
| 由来(天然) |
| |||||||||||||||||||||||||||||||||||||||||||||
| 由来(組換発現) | 生物種:  | |||||||||||||||||||||||||||||||||||||||||||||
| 緩衝液 | pH: 7.5 | |||||||||||||||||||||||||||||||||||||||||||||
| 緩衝液成分 |
| |||||||||||||||||||||||||||||||||||||||||||||
| 試料 | 包埋: NO / シャドウイング: NO / 染色: NO / 凍結: YES | |||||||||||||||||||||||||||||||||||||||||||||
| 試料支持 | グリッドの材料: GOLD / グリッドのタイプ: Quantifoil | |||||||||||||||||||||||||||||||||||||||||||||
| 急速凍結 | 凍結剤: ETHANE |
- 電子顕微鏡撮影
電子顕微鏡撮影
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
|---|---|
| 顕微鏡 | モデル: TFS KRIOS |
| 電子銃 | 電子線源:  FIELD EMISSION GUN / 加速電圧: 300 kV / 照射モード: FLOOD BEAM FIELD EMISSION GUN / 加速電圧: 300 kV / 照射モード: FLOOD BEAM |
| 電子レンズ | モード: BRIGHT FIELD / 倍率(補正後): 58139 X / 最大 デフォーカス(公称値): 3000 nm / 最小 デフォーカス(公称値): 1500 nm / Cs: 2.7 mm / アライメント法: COMA FREE |
| 試料ホルダ | 凍結剤: NITROGEN 試料ホルダーモデル: FEI TITAN KRIOS AUTOGRID HOLDER |
| 撮影 | 電子線照射量: 54 e/Å2 フィルム・検出器のモデル: GATAN K3 BIOQUANTUM (6k x 4k) |
| 電子光学装置 | エネルギーフィルター名称: GIF Bioquantum / エネルギーフィルタースリット幅: 20 eV |
- 解析
解析
| 画像処理 |
| |||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF補正 |
| |||||||||||||||||||||||||||||||||||
| 粒子像の選択 |
| |||||||||||||||||||||||||||||||||||
| 対称性 |
| |||||||||||||||||||||||||||||||||||
| 3次元再構成 |
| |||||||||||||||||||||||||||||||||||
| 原子モデル構築 | プロトコル: FLEXIBLE FIT / 空間: REAL | |||||||||||||||||||||||||||||||||||
| 精密化 | 最高解像度: 2.46 Å |
 ムービー
ムービー コントローラー
コントローラー













 PDBj
PDBj















































