[English] 日本語
 Yorodumi
Yorodumi- EMDB-29320: Structure of E138K/G140A/Q148R HIV-1 intasome with Dolutegravir bound -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of E138K/G140A/Q148R HIV-1 intasome with Dolutegravir bound | ||||||||||||||||||
 Map data Map data | Density modified and resampled map of E138K/G140A/Q148R HIV-1 intasome with drug Dolutegravir | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | Integrase / Nucleoprotein complex / Inhibitor / Drug resistance / VIRAL PROTEIN-DNA-INHIBITOR complex | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationDepolymerization of the Nuclear Lamina / Nuclear Envelope Breakdown / lamin binding / RHOD GTPase cycle / Initiation of Nuclear Envelope (NE) Reformation / RHOF GTPase cycle / nuclear inner membrane / RHOC GTPase cycle / RHOJ GTPase cycle / CDC42 GTPase cycle ...Depolymerization of the Nuclear Lamina / Nuclear Envelope Breakdown / lamin binding / RHOD GTPase cycle / Initiation of Nuclear Envelope (NE) Reformation / RHOF GTPase cycle / nuclear inner membrane / RHOC GTPase cycle / RHOJ GTPase cycle / CDC42 GTPase cycle / RHOG GTPase cycle / RHOA GTPase cycle / RAC2 GTPase cycle / RAC3 GTPase cycle / RAC1 GTPase cycle / HIV-1 retropepsin / symbiont-mediated activation of host apoptosis / retroviral ribonuclease H / exoribonuclease H / exoribonuclease H activity / DNA integration / viral genome integration into host DNA / establishment of integrated proviral latency / RNA-directed DNA polymerase / RNA stem-loop binding / viral penetration into host nucleus / host multivesicular body / RNA-directed DNA polymerase activity / RNA-DNA hybrid ribonuclease activity / Transferases; Transferring phosphorus-containing groups; Nucleotidyltransferases / nuclear envelope / host cell / viral nucleocapsid / DNA recombination / nuclear membrane / DNA-directed DNA polymerase / aspartic-type endopeptidase activity / Hydrolases; Acting on ester bonds / DNA-directed DNA polymerase activity / symbiont-mediated suppression of host gene expression / viral translational frameshifting / symbiont entry into host cell / lipid binding / host cell nucleus / host cell plasma membrane / virion membrane / structural molecule activity / proteolysis / DNA binding / zinc ion binding / membrane / nucleus / cytoplasm Similarity search - Function | ||||||||||||||||||
| Biological species |   Human immunodeficiency virus 1 Human immunodeficiency virus 1 | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.46 Å | ||||||||||||||||||
 Authors Authors | Shan ZL / Passos DO / Strutzenberg TS / Li M / Lyumkis D | ||||||||||||||||||
| Funding support |  United States, 5 items United States, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2023 Journal: Sci Adv / Year: 2023Title: Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants. Authors: Min Li / Dario Oliveira Passos / Zelin Shan / Steven J Smith / Qinfang Sun / Avik Biswas / Indrani Choudhuri / Timothy S Strutzenberg / Allan Haldane / Nanjie Deng / Zhaoyang Li / Xue Zhi ...Authors: Min Li / Dario Oliveira Passos / Zelin Shan / Steven J Smith / Qinfang Sun / Avik Biswas / Indrani Choudhuri / Timothy S Strutzenberg / Allan Haldane / Nanjie Deng / Zhaoyang Li / Xue Zhi Zhao / Lorenzo Briganti / Mamuka Kvaratskhelia / Terrence R Burke / Ronald M Levy / Stephen H Hughes / Robert Craigie / Dmitry Lyumkis /  Abstract: HIV-1 infection depends on the integration of viral DNA into host chromatin. Integration is mediated by the viral enzyme integrase and is blocked by integrase strand transfer inhibitors (INSTIs), ...HIV-1 infection depends on the integration of viral DNA into host chromatin. Integration is mediated by the viral enzyme integrase and is blocked by integrase strand transfer inhibitors (INSTIs), first-line antiretroviral therapeutics widely used in the clinic. Resistance to even the best INSTIs is a problem, and the mechanisms of resistance are poorly understood. Here, we analyze combinations of the mutations E138K, G140A/S, and Q148H/K/R, which confer resistance to INSTIs. The investigational drug 4d more effectively inhibited the mutants compared with the approved drug Dolutegravir (DTG). We present 11 new cryo-EM structures of drug-resistant HIV-1 intasomes bound to DTG or 4d, with better than 3-Å resolution. These structures, complemented with free energy simulations, virology, and enzymology, explain the mechanisms of DTG resistance involving E138K + G140A/S + Q148H/K/R and show why 4d maintains potency better than DTG. These data establish a foundation for further development of INSTIs that potently inhibit resistant forms in integrase. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29320.map.gz emd_29320.map.gz | 117.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29320-v30.xml emd-29320-v30.xml emd-29320.xml emd-29320.xml | 35.3 KB 35.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_29320.png emd_29320.png | 192.3 KB | ||
| Masks |  emd_29320_msk_1.map emd_29320_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-29320.cif.gz emd-29320.cif.gz | 8.9 KB | ||
| Others |  emd_29320_additional_1.map.gz emd_29320_additional_1.map.gz emd_29320_half_map_1.map.gz emd_29320_half_map_1.map.gz emd_29320_half_map_2.map.gz emd_29320_half_map_2.map.gz | 139 MB 139.1 MB 139.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29320 http://ftp.pdbj.org/pub/emdb/structures/EMD-29320 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29320 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29320 | HTTPS FTP |
-Related structure data
| Related structure data |  8fnoMC  8fn7C  8fndC  8fngC  8fnhC  8fnjC  8fnlC  8fnmC  8fnnC  8fnpC  8fnqC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29320.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29320.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Density modified and resampled map of E138K/G140A/Q148R HIV-1 intasome with drug Dolutegravir | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.38 Å | ||||||||||||||||||||||||||||||||||||
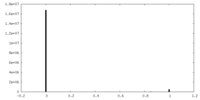
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_29320_msk_1.map emd_29320_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
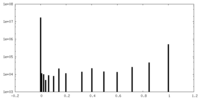
| Density Histograms |
-Additional map: Unsharpened map of E138K/G140A/Q148R HIV-1 intasome with drug...
| File | emd_29320_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map of E138K/G140A/Q148R HIV-1 intasome with drug Dolutegravir | ||||||||||||
| Projections & Slices |
| ||||||||||||
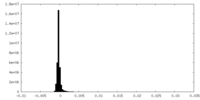
| Density Histograms |
-Half map: Half map1 of E138K/G140A/Q148R HIV-1 intasome with drug Dolutegravir
| File | emd_29320_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map1 of E138K/G140A/Q148R HIV-1 intasome with drug Dolutegravir | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map2 of E138K/G140A/Q148R HIV-1 intasome with drug Dolutegravir
| File | emd_29320_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map2 of E138K/G140A/Q148R HIV-1 intasome with drug Dolutegravir | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : E138K/G140A/Q148R HIV-1 intasome bound to dolutegravir
| Entire | Name: E138K/G140A/Q148R HIV-1 intasome bound to dolutegravir |
|---|---|
| Components |
|
-Supramolecule #1: E138K/G140A/Q148R HIV-1 intasome bound to dolutegravir
| Supramolecule | Name: E138K/G140A/Q148R HIV-1 intasome bound to dolutegravir type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 500 KDa |
-Macromolecule #1: Lamina-associated polypeptide 2, isoforms beta/gamma,Integrase
| Macromolecule | Name: Lamina-associated polypeptide 2, isoforms beta/gamma,Integrase type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO EC number: Transferases; Transferring phosphorus-containing groups; Nucleotidyltransferases |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 39.941512 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSHMPKRGRP AATEVKIPKP RGRPPLPAGT NSKGPPDFSS DEEREPTPVL GSGAAAAGQS RAAVGRKATK KTDGGGFLDG IDKAQEEHE KYHSNWRAMA SDFNLPPVVA KEIVASCDKC QLKGEAMHGQ VDCSPGIWQL DCTHLEGKVI LVAVHVASGY I EAEVIPAE ...String: GSHMPKRGRP AATEVKIPKP RGRPPLPAGT NSKGPPDFSS DEEREPTPVL GSGAAAAGQS RAAVGRKATK KTDGGGFLDG IDKAQEEHE KYHSNWRAMA SDFNLPPVVA KEIVASCDKC QLKGEAMHGQ VDCSPGIWQL DCTHLEGKVI LVAVHVASGY I EAEVIPAE TGQETAYFLL KLAGRWPVKT VHTDNGSNFT STTVKAACWW AGIKQKFAIP YNPQSRGVIE SMNKELKKII GQ VRDQAEH LKTAVQMAVF IHNFKRKGGI GGYSAGERIV DIIATDIQTK ELQKQITKIQ NFRVYYRDSR DPVWKGPAKL LWK GEGAVV IQDNSDIKVV PRRKAKIIRD YGKQMAGDDC VASRQDED UniProtKB: Lamina-associated polypeptide 2, isoforms beta/gamma, Gag-Pol polyprotein |
-Macromolecule #2: DNA (27-MER)
| Macromolecule | Name: DNA (27-MER) / type: dna / ID: 2 / Number of copies: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 8.188271 KDa |
| Sequence | String: (DA)(DC)(DT)(DG)(DC)(DT)(DA)(DG)(DA)(DG) (DA)(DT)(DT)(DT)(DT)(DC)(DC)(DC)(DG)(DC) (DC)(DC)(DA)(DC)(DG)(DC)(DT) |
-Macromolecule #3: DNA (25-MER)
| Macromolecule | Name: DNA (25-MER) / type: dna / ID: 3 / Number of copies: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 7.773023 KDa |
| Sequence | String: (DA)(DG)(DC)(DG)(DT)(DG)(DG)(DG)(DC)(DG) (DG)(DG)(DA)(DA)(DA)(DA)(DT)(DC)(DT)(DC) (DT)(DA)(DG)(DC)(DA) |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 4 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #5: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 5 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #6: (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4...
| Macromolecule | Name: (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide type: ligand / ID: 6 / Number of copies: 2 / Formula: DLU |
|---|---|
| Molecular weight | Theoretical: 419.379 Da |
| Chemical component information |  ChemComp-DLU: |
-Macromolecule #7: water
| Macromolecule | Name: water / type: ligand / ID: 7 / Number of copies: 890 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
| |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil / Material: GOLD / Support film - Material: GOLD / Support film - topology: HOLEY | |||||||||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 54.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 58139 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing #1
Image processing #1
+ Image processing #2
Image processing #2
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-8fno: |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)