[English] 日本語
 Yorodumi
Yorodumi- PDB-8fh4: Crystal structure of HPK1 kinase domain T165E,S171E phosphomimeti... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8fh4 | ||||||
|---|---|---|---|---|---|---|---|
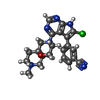
| Title | Crystal structure of HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with 3-[6-chloro-4-(9-methyl-1-oxa-4,9-diazaspiro[5.5]undec-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile | ||||||
 Components Components | Mitogen-activated protein kinase kinase kinase kinase 1 | ||||||
 Keywords Keywords | TRANSFERASE / inhibitor / hematopoietic / kinase | ||||||
| Function / homology |  Function and homology information Function and homology informationMAP kinase kinase kinase kinase activity / cellular response to phorbol 13-acetate 12-myristate / JNK cascade / peptidyl-serine phosphorylation / protein autophosphorylation / protein phosphorylation / cell population proliferation / non-specific serine/threonine protein kinase / positive regulation of MAPK cascade / intracellular signal transduction ...MAP kinase kinase kinase kinase activity / cellular response to phorbol 13-acetate 12-myristate / JNK cascade / peptidyl-serine phosphorylation / protein autophosphorylation / protein phosphorylation / cell population proliferation / non-specific serine/threonine protein kinase / positive regulation of MAPK cascade / intracellular signal transduction / protein serine kinase activity / protein serine/threonine kinase activity / ATP binding / membrane / cytoplasm Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.827 Å MOLECULAR REPLACEMENT / Resolution: 1.827 Å | ||||||
 Authors Authors | McTigue, M. / Johnson, E. / Cronin, C. | ||||||
| Funding support | 1items
| ||||||
 Citation Citation |  Journal: J.Med.Chem. / Year: 2023 Journal: J.Med.Chem. / Year: 2023Title: Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1. Authors: Gallego, R.A. / Bernier, L. / Chen, H. / Cho-Schultz, S. / Chung, L. / Collins, M. / Del Bel, M. / Elleraas, J. / Costa Jones, C. / Cronin, C.N. / Edwards, M. / Fang, X. / Fisher, T. / He, M. ...Authors: Gallego, R.A. / Bernier, L. / Chen, H. / Cho-Schultz, S. / Chung, L. / Collins, M. / Del Bel, M. / Elleraas, J. / Costa Jones, C. / Cronin, C.N. / Edwards, M. / Fang, X. / Fisher, T. / He, M. / Hoffman, J. / Huo, R. / Jalaie, M. / Johnson, E. / Johnson, T.W. / Kania, R.S. / Kraus, M. / Lafontaine, J. / Le, P. / Liu, T. / Maestre, M. / Matthews, J. / McTigue, M. / Miller, N. / Mu, Q. / Qin, X. / Ren, S. / Richardson, P. / Rohner, A. / Sach, N. / Shao, L. / Smith, G. / Su, R. / Sun, B. / Timofeevski, S. / Tran, P. / Wang, S. / Wang, W. / Zhou, R. / Zhu, J. / Nair, S.K. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8fh4.cif.gz 8fh4.cif.gz | 257.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8fh4.ent.gz pdb8fh4.ent.gz | 206.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8fh4.json.gz 8fh4.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/fh/8fh4 https://data.pdbj.org/pub/pdb/validation_reports/fh/8fh4 ftp://data.pdbj.org/pub/pdb/validation_reports/fh/8fh4 ftp://data.pdbj.org/pub/pdb/validation_reports/fh/8fh4 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  8fjzC  8fkoC  8fp1C  8fp3C  6ng0S S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| 3 | 
| ||||||||
| 4 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 37141.656 Da / Num. of mol.: 4 / Mutation: T165E, S171E Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: MAP4K1, HPK1 / Production host: Homo sapiens (human) / Gene: MAP4K1, HPK1 / Production host:  References: UniProt: Q92918, non-specific serine/threonine protein kinase #2: Chemical | ChemComp-XXF / ( #3: Chemical | ChemComp-SO4 / | #4: Water | ChemComp-HOH / | Has ligand of interest | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.2 Å3/Da / Density % sol: 44.18 % |
|---|---|
| Crystal grow | Temperature: 286.15 K / Method: vapor diffusion Details: 15 mg/mL protein + reservoir (0.1 M Tris, pH 8.0, 17.5% 1,6-hexanediol, 10 mM magnesium sulfate, 24 mM barium acetate) |
-Data collection
| Diffraction | Mean temperature: 180 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 17-ID / Wavelength: 1 Å / Beamline: 17-ID / Wavelength: 1 Å |
| Detector | Type: DECTRIS PILATUS3 6M / Detector: PIXEL / Date: Mar 2, 2016 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 1.83→69.56 Å / Num. obs: 92890 / % possible obs: 82.6 % / Redundancy: 1.8 % / CC1/2: 0.996 / Rmerge(I) obs: 0.03 / Rpim(I) all: 0.03 / Rrim(I) all: 0.042 / Net I/σ(I): 9.7 |
| Reflection shell | Resolution: 1.83→1.93 Å / Redundancy: 1.6 % / Rmerge(I) obs: 0.292 / Mean I/σ(I) obs: 1.9 / Num. unique obs: 4111 / CC1/2: 0.833 / Rpim(I) all: 0.292 / % possible all: 25 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB entry 6NG0 Resolution: 1.827→69.56 Å / Cor.coef. Fo:Fc: 0.946 / Cor.coef. Fo:Fc free: 0.939 / SU R Cruickshank DPI: 0.159 / Cross valid method: THROUGHOUT / SU R Blow DPI: 0.161 / SU Rfree Blow DPI: 0.137 / SU Rfree Cruickshank DPI: 0.138
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 31.69 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.22 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.827→69.56 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.83→1.85 Å
|
 Movie
Movie Controller
Controller


 PDBj
PDBj