[English] 日本語
 Yorodumi
Yorodumi- PDB-8dkb: Crystal Structure of human YEATS4 in complex with Pfizer small mo... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8dkb | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal Structure of human YEATS4 in complex with Pfizer small molecule compound 3b | ||||||
 Components Components | YEATS domain-containing protein 4 | ||||||
 Keywords Keywords | PROTEIN BINDING / histone binding / lysine-acetylated histone binding | ||||||
| Function / homology |  Function and homology information Function and homology informationhistone H3K18ac reader activity / histone H3K27ac reader activity / Activation of the TFAP2 (AP-2) family of transcription factors / regulation of double-strand break repair / NuA4 histone acetyltransferase complex / positive regulation of double-strand break repair via homologous recombination / structural constituent of cytoskeleton / nuclear matrix / nucleosome / mitotic cell cycle ...histone H3K18ac reader activity / histone H3K27ac reader activity / Activation of the TFAP2 (AP-2) family of transcription factors / regulation of double-strand break repair / NuA4 histone acetyltransferase complex / positive regulation of double-strand break repair via homologous recombination / structural constituent of cytoskeleton / nuclear matrix / nucleosome / mitotic cell cycle / HATs acetylate histones / nuclear membrane / histone binding / regulation of apoptotic process / regulation of cell cycle / chromatin remodeling / regulation of DNA-templated transcription / regulation of transcription by RNA polymerase II / positive regulation of DNA-templated transcription / nucleoplasm / nucleus Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.58 Å MOLECULAR REPLACEMENT / Resolution: 2.58 Å | ||||||
 Authors Authors | Dias, J.M. / Byrnes, L.J. / Varghese, A.H. | ||||||
| Funding support |  United States, 1items United States, 1items
| ||||||
 Citation Citation |  Journal: J.Med.Chem. / Year: 2023 Journal: J.Med.Chem. / Year: 2023Title: Discovery of High-Affinity Small-Molecule Binders of the Epigenetic Reader YEATS4. Authors: Londregan, A.T. / Aitmakhanova, K. / Bennett, J. / Byrnes, L.J. / Canterbury, D.P. / Cheng, X. / Christott, T. / Clemens, J. / Coffey, S.B. / Dias, J.M. / Dowling, M.S. / Farnie, G. / ...Authors: Londregan, A.T. / Aitmakhanova, K. / Bennett, J. / Byrnes, L.J. / Canterbury, D.P. / Cheng, X. / Christott, T. / Clemens, J. / Coffey, S.B. / Dias, J.M. / Dowling, M.S. / Farnie, G. / Fedorov, O. / Fennell, K.F. / Gamble, V. / Gileadi, C. / Giroud, C. / Harris, M.R. / Hollingshead, B.D. / Huber, K. / Korczynska, M. / Lapham, K. / Loria, P.M. / Narayanan, A. / Owen, D.R. / Raux, B. / Sahasrabudhe, P.V. / Ruggeri, R.B. / Saez, L.D. / Stock, I.A. / Thuma, B.A. / Tsai, A. / Varghese, A.E. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8dkb.cif.gz 8dkb.cif.gz | 456.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8dkb.ent.gz pdb8dkb.ent.gz | 380.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8dkb.json.gz 8dkb.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  8dkb_validation.pdf.gz 8dkb_validation.pdf.gz | 2 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  8dkb_full_validation.pdf.gz 8dkb_full_validation.pdf.gz | 2 MB | Display | |
| Data in XML |  8dkb_validation.xml.gz 8dkb_validation.xml.gz | 41.3 KB | Display | |
| Data in CIF |  8dkb_validation.cif.gz 8dkb_validation.cif.gz | 56.9 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/dk/8dkb https://data.pdbj.org/pub/pdb/validation_reports/dk/8dkb ftp://data.pdbj.org/pub/pdb/validation_reports/dk/8dkb ftp://data.pdbj.org/pub/pdb/validation_reports/dk/8dkb | HTTPS FTP |
-Related structure data
| Related structure data | 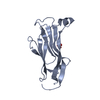 5vnaS S: Starting model for refinement |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
- Components
Components
| #1: Protein | Mass: 18230.885 Da / Num. of mol.: 8 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: YEATS4, GAS41 / Production host: Homo sapiens (human) / Gene: YEATS4, GAS41 / Production host:  #2: Chemical | ChemComp-SJI / #3: Water | ChemComp-HOH / | Has ligand of interest | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.35 Å3/Da / Density % sol: 47.57 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop Details: 0.1 M sodium citrate tribasic dihydrate (pH 5.6), 2% (v/v) tacsimate (pH 5.0), and 16% PEG 3350, 0.1M Ammonium sulfate |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 17-ID / Wavelength: 1 Å / Beamline: 17-ID / Wavelength: 1 Å |
| Detector | Type: DECTRIS PILATUS 6M / Detector: PIXEL / Date: Apr 10, 2019 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 2.58→103.32 Å / Num. obs: 26291 / % possible obs: 62 % / Redundancy: 3.4 % / CC1/2: 0.994 / Rsym value: 0.117 / Net I/σ(I): 8.7 |
| Reflection shell | Resolution: 2.582→2.803 Å / Redundancy: 3.4 % / Mean I/σ(I) obs: 1.3 / Num. unique obs: 1316 / CC1/2: 0.51 / Rsym value: 1.04 / % possible all: 14.2 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 5VNA Resolution: 2.58→41.3 Å / Cor.coef. Fo:Fc: 0.926 / Cor.coef. Fo:Fc free: 0.89 / Cross valid method: THROUGHOUT / σ(F): 0 / SU Rfree Blow DPI: 0.389
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 197.24 Å2 / Biso mean: 72.9 Å2 / Biso min: 15.6 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.31 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 2.58→41.3 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.58→2.73 Å / Rfactor Rfree error: 0 / Total num. of bins used: 50
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller


 PDBj
PDBj












