[English] 日本語
 Yorodumi
Yorodumi- PDB-8dhz: NMR Structure of Ac-hGal(17-30)NH2, an N-terminally acetylated fr... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8dhz | ||||||
|---|---|---|---|---|---|---|---|
| Title | NMR Structure of Ac-hGal(17-30)NH2, an N-terminally acetylated fragment of the C-terminus of human galanin | ||||||
 Components Components | Galanin | ||||||
 Keywords Keywords |  NEUROPEPTIDE / fragment NEUROPEPTIDE / fragment | ||||||
| Function / homology |  Function and homology information Function and homology information galanin receptor binding / type 1 galanin receptor binding / type 2 galanin receptor binding / type 3 galanin receptor binding / galanin receptor binding / type 1 galanin receptor binding / type 2 galanin receptor binding / type 3 galanin receptor binding /  galanin receptor activity / positive regulation of large conductance calcium-activated potassium channel activity / positive regulation of timing of catagen / positive regulation of cortisol secretion / regulation of glucocorticoid metabolic process / negative regulation of lymphocyte proliferation ... galanin receptor activity / positive regulation of large conductance calcium-activated potassium channel activity / positive regulation of timing of catagen / positive regulation of cortisol secretion / regulation of glucocorticoid metabolic process / negative regulation of lymphocyte proliferation ... galanin receptor binding / type 1 galanin receptor binding / type 2 galanin receptor binding / type 3 galanin receptor binding / galanin receptor binding / type 1 galanin receptor binding / type 2 galanin receptor binding / type 3 galanin receptor binding /  galanin receptor activity / positive regulation of large conductance calcium-activated potassium channel activity / positive regulation of timing of catagen / positive regulation of cortisol secretion / regulation of glucocorticoid metabolic process / negative regulation of lymphocyte proliferation / neuropeptide hormone activity / galanin receptor activity / positive regulation of large conductance calcium-activated potassium channel activity / positive regulation of timing of catagen / positive regulation of cortisol secretion / regulation of glucocorticoid metabolic process / negative regulation of lymphocyte proliferation / neuropeptide hormone activity /  feeding behavior / insulin secretion / response to immobilization stress / neuropeptide signaling pathway / protein kinase A signaling / cAMP-mediated signaling / Peptide ligand-binding receptors / feeding behavior / insulin secretion / response to immobilization stress / neuropeptide signaling pathway / protein kinase A signaling / cAMP-mediated signaling / Peptide ligand-binding receptors /  secretory granule / response to insulin / response to estrogen / G alpha (i) signalling events / response to xenobiotic stimulus / positive regulation of apoptotic process / neuronal cell body / positive regulation of transcription by RNA polymerase II / secretory granule / response to insulin / response to estrogen / G alpha (i) signalling events / response to xenobiotic stimulus / positive regulation of apoptotic process / neuronal cell body / positive regulation of transcription by RNA polymerase II /  extracellular space / extracellular region extracellular space / extracellular regionSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  SOLUTION NMR / SOLUTION NMR /  simulated annealing simulated annealing | ||||||
 Authors Authors | Kraichely, K.N. / Hendy, C.M. / Parnham, S. / Giuliano, M.W. | ||||||
| Funding support |  United States, 1items United States, 1items
| ||||||
 Citation Citation |  Journal: Biochem.Biophys.Res.Commun. / Year: 2022 Journal: Biochem.Biophys.Res.Commun. / Year: 2022Title: The neuropeptide galanin adopts an irregular secondary structure. Authors: Wilkinson, R.E. / Kraichely, K.N. / Hendy, C.M. / Buchanan, L.E. / Parnham, S. / Giuliano, M.W. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8dhz.cif.gz 8dhz.cif.gz | 100.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8dhz.ent.gz pdb8dhz.ent.gz | 70.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8dhz.json.gz 8dhz.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/dh/8dhz https://data.pdbj.org/pub/pdb/validation_reports/dh/8dhz ftp://data.pdbj.org/pub/pdb/validation_reports/dh/8dhz ftp://data.pdbj.org/pub/pdb/validation_reports/dh/8dhz | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  8dj4C C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
| Other databases |
|
- Links
Links
- Assembly
Assembly
| Deposited unit | 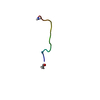
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide |  Mass: 1546.647 Da / Num. of mol.: 1 / Fragment: C-terminal residues, 49-62 / Source method: obtained synthetically / Source: (synth.)   Homo sapiens (human) / References: UniProt: P22466 Homo sapiens (human) / References: UniProt: P22466 |
|---|---|
| Has ligand of interest | N |
-Experimental details
-Experiment
| Experiment | Method:  SOLUTION NMR SOLUTION NMR | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details | Type: lyophilized powder Contents: 2 mM Ac-hGal(17-30)-NH2, 4.14 mM [U-2H] sodium acetate, 5.86 mM [U-2H] acetic acid, 95% H2O/5% D2O Details: peptide was suspended and dissolved into deuterated acetate buffer, which had been prepared in 95:5 H2O:D2O to within 0.02 units of pH 4.6. Label: Ac-hGal(17-30)-NH2 / Solvent system: 95% H2O/5% D2O | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| ||||||||||||||||
| Sample conditions | Ionic strength: sample was dissolved in 10 mM sodium acetate-d3/acetic acid-d4 buffer at pH 4.6; no additional additives were present. Not defined Label: 1 / pH: 4.6 / PH err: 0.02 / Pressure: 1 atm / Temperature: 298 K |
-NMR measurement
| NMR spectrometer | Type: Bruker AVANCE II / Manufacturer: Bruker / Model : AVANCE II / Field strength: 600 MHz : AVANCE II / Field strength: 600 MHz |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method:  simulated annealing / Software ordinal: 2 simulated annealing / Software ordinal: 2 | ||||||||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 1000 / Conformers submitted total number: 25 |
 Movie
Movie Controller
Controller





 PDBj
PDBj




