+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8bv1 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Peptide inhibitor P4 in complex with ASF1 histone chaperone | ||||||||||||
 Components Components |
| ||||||||||||
 Keywords Keywords | CHAPERONE / Inhibitor / ASF1 / Histone / protein-protein interaction | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationhistone chaperone activity / DNA replication-dependent chromatin assembly / muscle cell differentiation / Formation of Senescence-Associated Heterochromatin Foci (SAHF) / DNA repair-dependent chromatin remodeling / replication fork processing / osteoblast differentiation / nucleosome assembly / site of double-strand break / histone binding ...histone chaperone activity / DNA replication-dependent chromatin assembly / muscle cell differentiation / Formation of Senescence-Associated Heterochromatin Foci (SAHF) / DNA repair-dependent chromatin remodeling / replication fork processing / osteoblast differentiation / nucleosome assembly / site of double-strand break / histone binding / DNA repair / chromatin binding / chromatin / protein-containing complex / nucleoplasm / nucleus Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human)synthetic construct (others) | ||||||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.834 Å MOLECULAR REPLACEMENT / Resolution: 2.834 Å | ||||||||||||
 Authors Authors | Perrin, M.E. / Li, B. / Mbianda, J. / Ropars, V. / Legrand, P. / Douat, C. / Ochsenbein, F. / Guichard, G. | ||||||||||||
| Funding support |  France, 3items France, 3items
| ||||||||||||
 Citation Citation |  Journal: Chem.Commun.(Camb.) / Year: 2023 Journal: Chem.Commun.(Camb.) / Year: 2023Title: Unexpected binding modes of inhibitors to the histone chaperone ASF1 revealed by a foldamer scanning approach. Authors: Perrin, M.E. / Li, B. / Mbianda, J. / Bakail, M. / Andre, C. / Moal, G. / Legrand, P. / Ropars, V. / Douat, C. / Ochsenbein, F. / Guichard, G. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8bv1.cif.gz 8bv1.cif.gz | 418 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8bv1.ent.gz pdb8bv1.ent.gz | 351.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8bv1.json.gz 8bv1.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  8bv1_validation.pdf.gz 8bv1_validation.pdf.gz | 507.7 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  8bv1_full_validation.pdf.gz 8bv1_full_validation.pdf.gz | 513.5 KB | Display | |
| Data in XML |  8bv1_validation.xml.gz 8bv1_validation.xml.gz | 37.2 KB | Display | |
| Data in CIF |  8bv1_validation.cif.gz 8bv1_validation.cif.gz | 50.7 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/bv/8bv1 https://data.pdbj.org/pub/pdb/validation_reports/bv/8bv1 ftp://data.pdbj.org/pub/pdb/validation_reports/bv/8bv1 ftp://data.pdbj.org/pub/pdb/validation_reports/bv/8bv1 | HTTPS FTP |
-Related structure data
| Related structure data |  8cj1C  8cj2C  8cj3C  6f0hS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 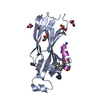
| ||||||||
| 2 | 
| ||||||||
| 3 | 
| ||||||||
| 4 | 
| ||||||||
| 5 | 
| ||||||||
| 6 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 17927.998 Da / Num. of mol.: 6 Source method: isolated from a genetically manipulated source Details: Histone chaperone that facilitates histone deposition and histone exchange and removal during nucleosome assembly and disassembly Source: (gene. exp.)  Homo sapiens (human) / Gene: ASF1A, CGI-98, HSPC146 / Plasmid: pETM30 / Production host: Homo sapiens (human) / Gene: ASF1A, CGI-98, HSPC146 / Plasmid: pETM30 / Production host:  #2: Protein/peptide | Mass: 1408.718 Da / Num. of mol.: 6 / Source method: obtained synthetically Details: Sequence of a short alpha-helical peptide known to bind ASF1 Source: (synth.) synthetic construct (others) #3: Chemical | ChemComp-GOL / #4: Water | ChemComp-HOH / | Has ligand of interest | N | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.48 Å3/Da / Density % sol: 50.1 % |
|---|---|
| Crystal grow | Temperature: 290.15 K / Method: vapor diffusion, sitting drop / pH: 8.5 / Details: 0.1 M Tris pH 8.5 30% (w/v) PEG 400 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SOLEIL SOLEIL  / Beamline: PROXIMA 1 / Wavelength: 0.97857 Å / Beamline: PROXIMA 1 / Wavelength: 0.97857 Å |
| Detector | Type: DECTRIS PILATUS 6M / Detector: PIXEL / Date: Feb 10, 2022 / Details: KB Mirrors |
| Radiation | Monochromator: 0.97857 / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.97857 Å / Relative weight: 1 |
| Reflection | Resolution: 2.834→49.372 Å / Num. obs: 56883 / % possible obs: 88.79 % / Redundancy: 14.27 % / CC1/2: 0.9979 / Rmerge(I) obs: 0.156 / Rpim(I) all: 0.043 / Rrim(I) all: 0.162 / Net I/σ(I): 13.13 |
| Reflection shell | Resolution: 2.834→2.907 Å / Redundancy: 13.97 % / Mean I/σ(I) obs: 1.446 / Num. unique obs: 1058 / CC1/2: 0.6066 / Rpim(I) all: 0.635 / % possible all: 22.49 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 6f0h Resolution: 2.834→29.79 Å / Cor.coef. Fo:Fc: 0.946 / Cor.coef. Fo:Fc free: 0.942 / SU R Cruickshank DPI: 0.355 / Cross valid method: THROUGHOUT / SU R Blow DPI: 0.332 / SU Rfree Blow DPI: 0.239 / SU Rfree Cruickshank DPI: 0.248
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 99.18 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.38 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.834→29.79 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.834→2.91 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller



 PDBj
PDBj









