[English] 日本語
 Yorodumi
Yorodumi- PDB-7zz3: Cryo-EM structure of "BC react" conformation of Lactococcus lacti... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7zz3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of "BC react" conformation of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | |||||||||
 Components Components | Pyruvate carboxylase | |||||||||
 Keywords Keywords | LIGASE / Tetramer / carboxylase / biotin | |||||||||
| Function / homology |  Function and homology information Function and homology informationpyruvate carboxylase / pyruvate carboxylase activity / pyruvate metabolic process / gluconeogenesis / ATP binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Lactococcus lactis (lactic acid bacteria) Lactococcus lactis (lactic acid bacteria) | |||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 2.41 Å | |||||||||
 Authors Authors | Lopez-Alonso, J.P. / Lazaro, M. / Gil, D. / Choi, P.H. / Tong, L. / Valle, M. | |||||||||
| Funding support |  France, France,  Spain, 2items Spain, 2items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase. Authors: Jorge Pedro López-Alonso / Melisa Lázaro / David Gil-Cartón / Philip H Choi / Alexandra Dodu / Liang Tong / Mikel Valle /   Abstract: Pyruvate carboxylase (PC) is a tetrameric enzyme that contains two active sites per subunit that catalyze two consecutive reactions. A mobile domain with an attached prosthetic biotin links both ...Pyruvate carboxylase (PC) is a tetrameric enzyme that contains two active sites per subunit that catalyze two consecutive reactions. A mobile domain with an attached prosthetic biotin links both reactions, an initial biotin carboxylation and the subsequent carboxyl transfer to pyruvate substrate to produce oxaloacetate. Reaction sites are at long distance, and there are several co-factors that play as allosteric regulators. Here, using cryoEM we explore the structure of active PC tetramers focusing on active sites and on the conformational space of the oligomers. The results capture the mobile domain at both active sites and expose catalytic steps of both reactions at high resolution, allowing the identification of substrates and products. The analysis of catalytically active PC tetramers reveals the role of certain motions during enzyme functioning, and the structural changes in the presence of additional cofactors expose the mechanism for allosteric regulation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7zz3.cif.gz 7zz3.cif.gz | 856.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7zz3.ent.gz pdb7zz3.ent.gz | 686.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7zz3.json.gz 7zz3.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  7zz3_validation.pdf.gz 7zz3_validation.pdf.gz | 1.7 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  7zz3_full_validation.pdf.gz 7zz3_full_validation.pdf.gz | 1.8 MB | Display | |
| Data in XML |  7zz3_validation.xml.gz 7zz3_validation.xml.gz | 121.4 KB | Display | |
| Data in CIF |  7zz3_validation.cif.gz 7zz3_validation.cif.gz | 184.2 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/zz/7zz3 https://data.pdbj.org/pub/pdb/validation_reports/zz/7zz3 ftp://data.pdbj.org/pub/pdb/validation_reports/zz/7zz3 ftp://data.pdbj.org/pub/pdb/validation_reports/zz/7zz3 | HTTPS FTP |
-Related structure data
| Related structure data |  15033MC  7zyyC  7zyzC  7zz0C  7zz1C  7zz2C  7zz4C  7zz5C  7zz6C  7zz8C C: citing same article ( M: map data used to model this data |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Protein , 1 types, 4 molecules ABCD
| #1: Protein | Mass: 127349.578 Da / Num. of mol.: 4 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Lactococcus lactis (lactic acid bacteria) Lactococcus lactis (lactic acid bacteria)Gene: BW154_09950 / Production host:  |
|---|
-Non-polymers , 9 types, 192 molecules 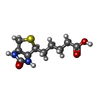
















| #2: Chemical | ChemComp-BTN / | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| #3: Chemical | ChemComp-MG / #4: Chemical | ChemComp-MN / #5: Chemical | ChemComp-ATP / | #6: Chemical | ChemComp-ACO / #7: Chemical | ChemComp-PYR / #8: Chemical | ChemComp-BCT / | #9: Chemical | #10: Water | ChemComp-HOH / | |
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: 3D ARRAY / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Pyruvate carboxylase with acetyl coenzyme A / Type: ORGANELLE OR CELLULAR COMPONENT / Entity ID: #1 / Source: RECOMBINANT | |||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Source (natural) | Organism:  Lactococcus lactis (lactic acid bacteria) Lactococcus lactis (lactic acid bacteria) | |||||||||||||||||||||||||||||||||||||||||||||
| Source (recombinant) | Organism:  | |||||||||||||||||||||||||||||||||||||||||||||
| Buffer solution | pH: 7.6 | |||||||||||||||||||||||||||||||||||||||||||||
| Buffer component |
| |||||||||||||||||||||||||||||||||||||||||||||
| Specimen | Conc.: 0.05 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES | |||||||||||||||||||||||||||||||||||||||||||||
| Specimen support | Grid material: COPPER / Grid mesh size: 300 divisions/in. / Grid type: Quantifoil R2/1 | |||||||||||||||||||||||||||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 81000 X / Nominal defocus max: 2400 nm / Nominal defocus min: 900 nm / Cs: 2.7 mm |
| Image recording | Average exposure time: 3.99 sec. / Electron dose: 48 e/Å2 / Detector mode: COUNTING / Film or detector model: GATAN K3 (6k x 4k) / Num. of grids imaged: 1 / Num. of real images: 10518 |
- Processing
Processing
| Software | Name: PHENIX / Version: 1.20.1_4487: / Classification: refinement | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software |
| ||||||||||||||||||||||||||||||||||||
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 4578464 | ||||||||||||||||||||||||||||||||||||
| Symmetry | Point symmetry: C1 (asymmetric) | ||||||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 2.41 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 128647 / Num. of class averages: 1 / Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||
| Atomic model building | Protocol: FLEXIBLE FIT / Space: REAL | ||||||||||||||||||||||||||||||||||||
| Atomic model building | PDB-ID: 5VYZ Accession code: 5VYZ / Source name: PDB / Type: experimental model | ||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller











 PDBj
PDBj



















