[English] 日本語
 Yorodumi
Yorodumi- EMDB-15034: Cryo-EM structure of "BC closed" conformation of Lactococcus lact... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of "BC closed" conformation of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Tetramer / carboxylase / biotin / inhibitor / LIGASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationpyruvate carboxylase / pyruvate carboxylase activity / pyruvate metabolic process / gluconeogenesis / ATP binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Lactococcus lactis (lactic acid bacteria) Lactococcus lactis (lactic acid bacteria) | |||||||||
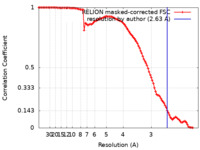
| Method | single particle reconstruction / cryo EM / Resolution: 2.63 Å | |||||||||
 Authors Authors | Lopez-Alonso JP / Lazaro M / Gil D / Choi PH / Tong L / Valle M | |||||||||
| Funding support |  Spain, 2 items Spain, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase. Authors: Jorge Pedro López-Alonso / Melisa Lázaro / David Gil-Cartón / Philip H Choi / Alexandra Dodu / Liang Tong / Mikel Valle /   Abstract: Pyruvate carboxylase (PC) is a tetrameric enzyme that contains two active sites per subunit that catalyze two consecutive reactions. A mobile domain with an attached prosthetic biotin links both ...Pyruvate carboxylase (PC) is a tetrameric enzyme that contains two active sites per subunit that catalyze two consecutive reactions. A mobile domain with an attached prosthetic biotin links both reactions, an initial biotin carboxylation and the subsequent carboxyl transfer to pyruvate substrate to produce oxaloacetate. Reaction sites are at long distance, and there are several co-factors that play as allosteric regulators. Here, using cryoEM we explore the structure of active PC tetramers focusing on active sites and on the conformational space of the oligomers. The results capture the mobile domain at both active sites and expose catalytic steps of both reactions at high resolution, allowing the identification of substrates and products. The analysis of catalytically active PC tetramers reveals the role of certain motions during enzyme functioning, and the structural changes in the presence of additional cofactors expose the mechanism for allosteric regulation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15034.map.gz emd_15034.map.gz | 857.3 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15034-v30.xml emd-15034-v30.xml emd-15034.xml emd-15034.xml | 21.4 KB 21.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15034_fsc.xml emd_15034_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_15034.png emd_15034.png | 88 KB | ||
| Masks |  emd_15034_msk_1.map emd_15034_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-15034.cif.gz emd-15034.cif.gz | 7 KB | ||
| Others |  emd_15034_half_map_1.map.gz emd_15034_half_map_1.map.gz emd_15034_half_map_2.map.gz emd_15034_half_map_2.map.gz | 81 MB 81 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15034 http://ftp.pdbj.org/pub/emdb/structures/EMD-15034 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15034 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15034 | HTTPS FTP |
-Related structure data
| Related structure data |  7zz4MC  7zyyC  7zyzC  7zz0C  7zz1C  7zz2C  7zz3C  7zz5C  7zz6C  7zz8C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15034.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15034.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||
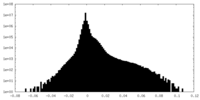
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15034_msk_1.map emd_15034_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
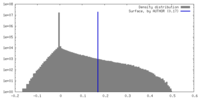
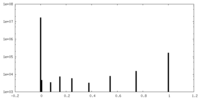
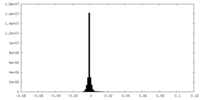
| Density Histograms |
-Half map: #1
| File | emd_15034_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_15034_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Pyruvate carboxylase with acetyl coenzyme A
| Entire | Name: Pyruvate carboxylase with acetyl coenzyme A |
|---|---|
| Components |
|
-Supramolecule #1: Pyruvate carboxylase with acetyl coenzyme A
| Supramolecule | Name: Pyruvate carboxylase with acetyl coenzyme A / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Lactococcus lactis (lactic acid bacteria) Lactococcus lactis (lactic acid bacteria) |
-Macromolecule #1: Pyruvate carboxylase
| Macromolecule | Name: Pyruvate carboxylase / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: pyruvate carboxylase |
|---|---|
| Source (natural) | Organism:  Lactococcus lactis (lactic acid bacteria) Lactococcus lactis (lactic acid bacteria) |
| Molecular weight | Theoretical: 127.349578 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: VPRGSHMKKL LVANRGEIAV RVFRACNELG LSTVAVYARE DEYSVHRFKA DESYLIGQGK KPIDAYLDID DIIRVALESG ADAIHPGYG LLSENLEFAT KVRAAGLVFV GPELHHLDIF GDKIKAKAAA DEAKVPGIPG TNGAVDIDGA LEFAKTYGYP V MIKAALGG ...String: VPRGSHMKKL LVANRGEIAV RVFRACNELG LSTVAVYARE DEYSVHRFKA DESYLIGQGK KPIDAYLDID DIIRVALESG ADAIHPGYG LLSENLEFAT KVRAAGLVFV GPELHHLDIF GDKIKAKAAA DEAKVPGIPG TNGAVDIDGA LEFAKTYGYP V MIKAALGG GGRGMRVARN DAEMHDGYAR AKSEAIGAFG SGEIYVEKYI ENPKHIEVQI LGDRHGNIIH LHERDCSVQR RN QKVIEIA PAVGLSPDFR NEICEAAVKL CKNVGYVNAG TVEFLVKDDK FYFIEVNPRV QVEHTITELI TGVDIVQAQI LIA QGKDLH REIGLPAQSE IPLLGSAIQC RITTEDPQNG FLPDTGKIDT YRSPGGFGIR LDVGNAYAGY EVTPYFDSLL VKVC TFANE FSDSVRKMDR VLHEFRIRGV KTNIPFLINV IANENFTSGQ ATTTFIDNTP SLFNFPRLRD RGTKTLHYLS MITVN GFPG IENTEKRHFE EPRQPLLNLE KKKTAKNILD EQGADAVVDY VKNTKEVLLT DTTLRDAHQS LLATRLRLQD MKGIAQ AID QGLPELFSAE MWGGATFDVA YRFLNESPWY RLRKLRKLMP NTMFQMLFRG SNAVGYQNYP DNVIEEFIRV AAHEGID VF RIFDSLNWLP QMEKSIQAVR DNGKIAEATI CYTGDILDPS RPKYNIQYYK DLAKELEATG AHILAV(KCX)DMA GLLK PQAAY RLISELKDTV DLPIHLHTHD TSGNGIITYS GATQAGVDII DVATASLAGG TSQPSMQSIY YALEHGPRHA SINVK NAEQ IDHYWEDVRK YYAPFEAGIT SPQTEVYMHE MPGGQYTNLK SQAAAVGLGH RFDEIKQMYR KVNMMFGDII KVTPSS KVV GDMALFMIQN DLTEEDVYAR GNELNFPESV VSFFRGDLGQ PVGGFPEKLQ KIIVKDKAVI TDRPGLHAEK VDFETVK AD LEQKIGYEPG DHEVISYIMY PQVFLDYQKM QREFGAVTLL DTPTFLHGMR LNEKIEVQIE KGKTLSIRLD EIGEPDLA G NRVLFFNLNG QRREVVINDQ SVQAQVVAKR KAETGNPNQI GATMPGSVLE ILVKAGDKVQ KGQALMVTEA MKMETTIEA PFDGEIVDLH VVKGEAIQTQ DLLIEIN UniProtKB: Pyruvate carboxylase |
-Macromolecule #2: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 1 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #4: ACETYL COENZYME *A
| Macromolecule | Name: ACETYL COENZYME *A / type: ligand / ID: 4 / Number of copies: 1 / Formula: ACO |
|---|---|
| Molecular weight | Theoretical: 809.571 Da |
| Chemical component information |  ChemComp-ACO: |
-Macromolecule #5: water
| Macromolecule | Name: water / type: ligand / ID: 5 / Number of copies: 11 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | 3D array |
- Sample preparation
Sample preparation
| Concentration | 0.05 mg/mL | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.6 Component:
| |||||||||||||||||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE | |||||||||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 10518 / Average exposure time: 3.99 sec. / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.9 µm / Nominal magnification: 81000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)