[English] 日本語
 Yorodumi
Yorodumi- PDB-7zjq: Human TEAD3 in complex with 1-Cyclopentyl-1H-pyrazolo[3,4-b]pyrid... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7zjq | ||||||
|---|---|---|---|---|---|---|---|
| Title | Human TEAD3 in complex with 1-Cyclopentyl-1H-pyrazolo[3,4-b]pyridine-5-carboxylic acid | ||||||
 Components Components | (Transcriptional enhancer factor TEF-5) x 2 | ||||||
 Keywords Keywords | TRANSCRIPTION / IMMUNOGLOBULIN-LIKE FOLD / ACTIVATOR / DISEASE MUTATION / DNA-BINDING / NUCLEUS / PHOSPHOPROTEIN / TRANSCRIPTION REGULATION / TRANSCRIPTION-PROTEIN BINDING COMPLEX | ||||||
| Function / homology |  Function and homology information Function and homology informationasymmetric neuroblast division / RUNX3 regulates YAP1-mediated transcription / YAP1- and WWTR1 (TAZ)-stimulated gene expression / hippo signaling / embryonic organ development / female pregnancy / transcription regulator complex / RNA polymerase II-specific DNA-binding transcription factor binding / DNA-binding transcription factor activity, RNA polymerase II-specific / RNA polymerase II cis-regulatory region sequence-specific DNA binding ...asymmetric neuroblast division / RUNX3 regulates YAP1-mediated transcription / YAP1- and WWTR1 (TAZ)-stimulated gene expression / hippo signaling / embryonic organ development / female pregnancy / transcription regulator complex / RNA polymerase II-specific DNA-binding transcription factor binding / DNA-binding transcription factor activity, RNA polymerase II-specific / RNA polymerase II cis-regulatory region sequence-specific DNA binding / DNA-binding transcription factor activity / regulation of transcription by RNA polymerase II / chromatin / positive regulation of transcription by RNA polymerase II / nucleoplasm Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.095 Å MOLECULAR REPLACEMENT / Resolution: 2.095 Å | ||||||
 Authors Authors | Musil, D. / Sousa, C.M. | ||||||
| Funding support | 1items
| ||||||
 Citation Citation |  Journal: J.Med.Chem. / Year: 2022 Journal: J.Med.Chem. / Year: 2022Title: Optimization of TEAD P-Site Binding Fragment Hit into In Vivo Active Lead MSC-4106 . Authors: Heinrich, T. / Peterson, C. / Schneider, R. / Garg, S. / Schwarz, D. / Gunera, J. / Seshire, A. / Kotzner, L. / Schlesiger, S. / Musil, D. / Schilke, H. / Doerfel, B. / Diehl, P. / Bopple, P. ...Authors: Heinrich, T. / Peterson, C. / Schneider, R. / Garg, S. / Schwarz, D. / Gunera, J. / Seshire, A. / Kotzner, L. / Schlesiger, S. / Musil, D. / Schilke, H. / Doerfel, B. / Diehl, P. / Bopple, P. / Lemos, A.R. / Sousa, P.M.F. / Freire, F. / Bandeiras, T.M. / Carswell, E. / Pearson, N. / Sirohi, S. / Hooker, M. / Trivier, E. / Broome, R. / Balsiger, A. / Crowden, A. / Dillon, C. / Wienke, D. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7zjq.cif.gz 7zjq.cif.gz | 356.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7zjq.ent.gz pdb7zjq.ent.gz | 292.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7zjq.json.gz 7zjq.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/zj/7zjq https://data.pdbj.org/pub/pdb/validation_reports/zj/7zjq ftp://data.pdbj.org/pub/pdb/validation_reports/zj/7zjq ftp://data.pdbj.org/pub/pdb/validation_reports/zj/7zjq | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  7zjpC  5emwS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 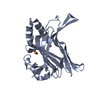
| ||||||||
| 2 | 
| ||||||||
| 3 | 
| ||||||||
| 4 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 25324.789 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: TEAD3, TEAD5, TEF5 / Production host: Homo sapiens (human) / Gene: TEAD3, TEAD5, TEF5 / Production host:  | ||||||
|---|---|---|---|---|---|---|---|
| #2: Protein | Mass: 25308.789 Da / Num. of mol.: 3 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: TEAD3, TEAD5, TEF5 / Production host: Homo sapiens (human) / Gene: TEAD3, TEAD5, TEF5 / Production host:  #3: Chemical | ChemComp-JIK / #4: Water | ChemComp-HOH / | Has ligand of interest | Y | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.04 Å3/Da / Density % sol: 59.47 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop / pH: 4.5 Details: 100 mM Sodium Acetate, 100 mM Calcium acetate,, 10% PEG 4000 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SLS SLS  / Beamline: X06SA / Wavelength: 0.9998 Å / Beamline: X06SA / Wavelength: 0.9998 Å |
| Detector | Type: DECTRIS PILATUS 6M-F / Detector: PIXEL / Date: Nov 12, 2016 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9998 Å / Relative weight: 1 |
| Reflection | Resolution: 2.095→47.771 Å / Num. obs: 72638 / % possible obs: 99.2 % / Redundancy: 6.7 % / CC1/2: 1 / Rrim(I) all: 0.095 / Net I/σ(I): 13.6 |
| Reflection shell | Resolution: 2.095→2.22 Å / Num. unique obs: 11388 / CC1/2: 0.04 / % possible all: 97.6 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 5EMW Resolution: 2.095→47.77 Å / Cor.coef. Fo:Fc: 0.923 / Cor.coef. Fo:Fc free: 0.917 / SU R Cruickshank DPI: 0.226 / Cross valid method: THROUGHOUT / σ(F): 0 / SU R Blow DPI: 0.22 / SU Rfree Blow DPI: 0.185 / SU Rfree Cruickshank DPI: 0.19
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 243.9 Å2 / Biso mean: 92.75 Å2 / Biso min: 49.53 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.89 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 2.095→47.77 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.1→2.12 Å / Rfactor Rfree error: 0 / Total num. of bins used: 51
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller


 PDBj
PDBj