+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7zj5 | ||||||
|---|---|---|---|---|---|---|---|
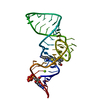
| Title | Unbound state of a brocolli-pepper aptamer FRET tile. | ||||||
 Components Components | brocolli-pepper aptamer | ||||||
 Keywords Keywords | RNA / origami / aptamer / fret | ||||||
| Function / homology | : / RNA / RNA (> 10) / RNA (> 100) Function and homology information Function and homology information | ||||||
| Biological species | synthetic construct (others) | ||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 4.55 Å | ||||||
 Authors Authors | McRae, E.K.S. / Vallina, N.S. / Hansen, B.K. / Boussebayle, A. / Andersen, E.S. | ||||||
| Funding support |  Denmark, 1items Denmark, 1items
| ||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structure determination of Pepper-Broccoli FRET pair by RNA origami scaffolding Authors: McRae, E.K.S. / Vallina, N.S. / Hansen, B.K. / Boussebayle, A. / Andersen, E.S. #1:  Journal: Acta Crystallogr D Struct Biol / Year: 2018 Journal: Acta Crystallogr D Struct Biol / Year: 2018Title: Real-space refinement in PHENIX for cryo-EM and crystallography. Authors: Afonine, P.V. / Poon, B.K. / Read, R.J. / Sobolev, O.V. / Terwilliger, T.C. / Urzhumtsev, A. / Adams, P.D. #2:  Journal: Acta Crystallogr D Struct Biol / Year: 2018 Journal: Acta Crystallogr D Struct Biol / Year: 2018Title: ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Authors: Croll, T.I. #3:  Journal: Nat Methods / Year: 2017 Journal: Nat Methods / Year: 2017Title: cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Authors: Punjani, A. / Rubinstein, J.L. / Fleet, D.J. / Brubaker, M.A. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7zj5.cif.gz 7zj5.cif.gz | 269.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7zj5.ent.gz pdb7zj5.ent.gz | Display |  PDB format PDB format | |
| PDBx/mmJSON format |  7zj5.json.gz 7zj5.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  7zj5_validation.pdf.gz 7zj5_validation.pdf.gz | 1.1 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  7zj5_full_validation.pdf.gz 7zj5_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  7zj5_validation.xml.gz 7zj5_validation.xml.gz | 22.1 KB | Display | |
| Data in CIF |  7zj5_validation.cif.gz 7zj5_validation.cif.gz | 32.4 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/zj/7zj5 https://data.pdbj.org/pub/pdb/validation_reports/zj/7zj5 ftp://data.pdbj.org/pub/pdb/validation_reports/zj/7zj5 ftp://data.pdbj.org/pub/pdb/validation_reports/zj/7zj5 | HTTPS FTP |
-Related structure data
| Related structure data |  14741MC  7zj4C M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: RNA chain | Mass: 120700.953 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.) synthetic construct (others) / Production host: synthetic construct (others) |
|---|---|
| #2: Chemical | ChemComp-K / |
| Has ligand of interest | N |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Unbound state of a brocolli-pepper aptamer FRET tile / Type: COMPLEX / Entity ID: #1 / Source: RECOMBINANT |
|---|---|
| Molecular weight | Value: 0.12 MDa / Experimental value: NO |
| Source (natural) | Organism: synthetic construct (others) |
| Source (recombinant) | Organism: synthetic construct (others) |
| Buffer solution | pH: 7.5 Details: 40mM HEPES pH 7.5, 5mM MgCl2, 50mM KCl. Filtered through 0.22 um filter. |
| Specimen | Conc.: 2.5 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES Details: Sample was purified by size exclusion chromatography. |
| Vitrification | Instrument: LEICA EM GP / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 288 K Details: 3 uL sample, blotted onto double layer of whatman filter paper for 6 seconds. |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal defocus max: 2000 nm / Nominal defocus min: 700 nm / Cs: 2.7 mm |
| Image recording | Electron dose: 60 e/Å2 / Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Num. of grids imaged: 1 / Num. of real images: 1605 Details: Collected with a calibrated pixel size of 0.647 Angstrom |
| EM imaging optics | Energyfilter name: GIF Bioquantum / Energyfilter slit width: 20 eV |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software |
| ||||||||||||||||||||||||||||||||||||
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 478981 Details: Picked from templates generated from an ab initio model | ||||||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 4.55 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 51278 / Algorithm: BACK PROJECTION Details: Local refinement was performed in cryoSPARC using a mask that covers the entire particle volume Num. of class averages: 1 / Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||
| Atomic model building | Protocol: FLEXIBLE FIT / Space: REAL | ||||||||||||||||||||||||||||||||||||
| Atomic model building |
| ||||||||||||||||||||||||||||||||||||
| Refinement | Cross valid method: NONE | ||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller




 PDBj
PDBj