+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Unbound state of a brocolli-pepper aptamer FRET tile | |||||||||
 Map data Map data | Map sharpened with a b-factor of 160. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RNA / origami / aptamer / fret | |||||||||
| Biological species | synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.55 Å | |||||||||
 Authors Authors | McRae EKS / Vallina NS / Hansen BK / Boussebayle A / Andersen ES | |||||||||
| Funding support |  Denmark, 1 items Denmark, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structure determination of Pepper-Broccoli FRET pair by RNA origami scaffolding Authors: McRae EKS / Vallina NS / Hansen BK / Boussebayle A / Andersen ES #1:  Journal: Acta Crystallogr D Struct Biol / Year: 2018 Journal: Acta Crystallogr D Struct Biol / Year: 2018Title: Real-space refinement in PHENIX for cryo-EM and crystallography. Authors: Afonine PV / Poon BK / Read RJ / Sobolev OV / Terwilliger TC / Urzhumtsev A / Adams PD #2:  Journal: Acta Crystallogr D Struct Biol / Year: 2018 Journal: Acta Crystallogr D Struct Biol / Year: 2018Title: ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Authors: Croll TI #3:  Journal: Nat Methods / Year: 2017 Journal: Nat Methods / Year: 2017Title: cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Authors: Punjani A / Rubinstein JL / Fleet DJ / Brubaker MA | |||||||||
| History |
|
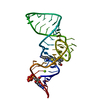
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14741.map.gz emd_14741.map.gz | 36.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14741-v30.xml emd-14741-v30.xml emd-14741.xml emd-14741.xml | 21.9 KB 21.9 KB | Display Display |  EMDB header EMDB header |
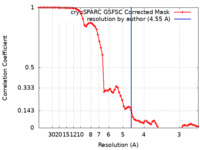
| FSC (resolution estimation) |  emd_14741_fsc.xml emd_14741_fsc.xml | 7.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_14741.png emd_14741.png | 28 KB | ||
| Masks |  emd_14741_msk_1.map emd_14741_msk_1.map | 38.4 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-14741.cif.gz emd-14741.cif.gz | 6 KB | ||
| Others |  emd_14741_additional_1.map.gz emd_14741_additional_1.map.gz emd_14741_half_map_1.map.gz emd_14741_half_map_1.map.gz emd_14741_half_map_2.map.gz emd_14741_half_map_2.map.gz | 19.2 MB 35.7 MB 35.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14741 http://ftp.pdbj.org/pub/emdb/structures/EMD-14741 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14741 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14741 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14741.map.gz / Format: CCP4 / Size: 38.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14741.map.gz / Format: CCP4 / Size: 38.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map sharpened with a b-factor of 160. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.29 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_14741_msk_1.map emd_14741_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Unsharpened map
| File | emd_14741_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A
| File | emd_14741_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B
| File | emd_14741_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Unbound state of a brocolli-pepper aptamer FRET tile
| Entire | Name: Unbound state of a brocolli-pepper aptamer FRET tile |
|---|---|
| Components |
|
-Supramolecule #1: Unbound state of a brocolli-pepper aptamer FRET tile
| Supramolecule | Name: Unbound state of a brocolli-pepper aptamer FRET tile / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 120 KDa |
-Macromolecule #1: brocolli-pepper aptamer
| Macromolecule | Name: brocolli-pepper aptamer / type: rna / ID: 1 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 120.700953 KDa |
| Sequence | String: GGAUACGUCU ACGCUCAGUG ACGGACUCUC UUCGGAGAGU CUGACAUCCG AACCAUACAC GGAUGUGCCU CGCCGAACAG UCUACGGCG AGCUUAAGCG CUGGGGACGC CCAACGCAUC ACAAAGACUG AGUGAUGAAC CAGAAGUAUG GACUGGUUGC G UUGGUGGA ...String: GGAUACGUCU ACGCUCAGUG ACGGACUCUC UUCGGAGAGU CUGACAUCCG AACCAUACAC GGAUGUGCCU CGCCGAACAG UCUACGGCG AGCUUAAGCG CUGGGGACGC CCAACGCAUC ACAAAGACUG AGUGAUGAAC CAGAAGUAUG GACUGGUUGC G UUGGUGGA GACGGUCGGG UCCAGUUCGC UGUCGAGUAG AGUGUGGGCU CCAUCGACGC CGCUUUAAGG UCCCCAAUCG UG GCGUGUC GGCCUGCUUC GGCAGGCACU GGCGCCGGGA CCUUGAAGAG AUGAGAUUUC GAUCUCAUCU UUGGGUGUCU CUG GUGCUU GAGGGCCCUG UGUUCGCACA GGGCCGCUCA CUGGGUGUGG ACGUAUCC |
-Macromolecule #2: POTASSIUM ION
| Macromolecule | Name: POTASSIUM ION / type: ligand / ID: 2 / Number of copies: 1 / Formula: K |
|---|---|
| Molecular weight | Theoretical: 39.098 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 40mM HEPES pH 7.5, 5mM MgCl2, 50mM KCl. Filtered through 0.22 um filter. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 288 K / Instrument: LEICA EM GP Details: 3 uL sample, blotted onto double layer of whatman filter paper for 6 seconds.. |
| Details | Sample was purified by size exclusion chromatography. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 1605 / Average electron dose: 60.0 e/Å2 Details: Collected with a calibrated pixel size of 0.647 Angstrom |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.7000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)