[English] 日本語
 Yorodumi
Yorodumi- PDB-7wsm: Cryo-EM structure of human glucose transporter GLUT4 bound to cyt... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7wsm | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human glucose transporter GLUT4 bound to cytochalasin B in lipid nanodiscs | |||||||||||||||||||||||||||
 Components Components | Solute carrier family 2, facilitated glucose transporter member 4 | |||||||||||||||||||||||||||
 Keywords Keywords | TRANSPORT PROTEIN / glucose transporter / GLUT4 / diabetes / cytochalasin B | |||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationD-glucose uniporter activity / regulation of synaptic vesicle budding from presynaptic endocytic zone membrane / positive regulation of brain-derived neurotrophic factor receptor signaling pathway / white fat cell proliferation / amylopectin biosynthetic process / dehydroascorbic acid transport / glucose import in response to insulin stimulus / Cellular hexose transport / D-glucose transmembrane transporter activity / insulin-responsive compartment ...D-glucose uniporter activity / regulation of synaptic vesicle budding from presynaptic endocytic zone membrane / positive regulation of brain-derived neurotrophic factor receptor signaling pathway / white fat cell proliferation / amylopectin biosynthetic process / dehydroascorbic acid transport / glucose import in response to insulin stimulus / Cellular hexose transport / D-glucose transmembrane transporter activity / insulin-responsive compartment / D-glucose transmembrane transport / trans-Golgi network transport vesicle / short-term memory / cellular response to osmotic stress / vesicle membrane / clathrin-coated vesicle / : / long-term memory / brown fat cell differentiation / transport across blood-brain barrier / multivesicular body / clathrin-coated pit / endomembrane system / T-tubule / cytoplasmic vesicle membrane / sarcoplasmic reticulum / Translocation of SLC2A4 (GLUT4) to the plasma membrane / trans-Golgi network / sarcolemma / Transcriptional regulation of white adipocyte differentiation / cellular response to insulin stimulus / cellular response to tumor necrosis factor / glucose homeostasis / presynapse / response to ethanol / cellular response to hypoxia / carbohydrate metabolic process / learning or memory / membrane raft / external side of plasma membrane / perinuclear region of cytoplasm / extracellular exosome / membrane / plasma membrane / cytosol Similarity search - Function | |||||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.25 Å | |||||||||||||||||||||||||||
 Authors Authors | Yuan, Y. / Kong, F. / Xu, H. / Zhu, A. / Yan, N. / Yan, C. | |||||||||||||||||||||||||||
| Funding support |  China, 2items China, 2items
| |||||||||||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Cryo-EM structure of human glucose transporter GLUT4. Authors: Yafei Yuan / Fang Kong / Hanwen Xu / Angqi Zhu / Nieng Yan / Chuangye Yan /   Abstract: GLUT4 is the primary glucose transporter in adipose and skeletal muscle tissues. Its cellular trafficking is regulated by insulin signaling. Failed or reduced plasma membrane localization of GLUT4 is ...GLUT4 is the primary glucose transporter in adipose and skeletal muscle tissues. Its cellular trafficking is regulated by insulin signaling. Failed or reduced plasma membrane localization of GLUT4 is associated with diabetes. Here, we report the cryo-EM structures of human GLUT4 bound to a small molecule inhibitor cytochalasin B (CCB) at resolutions of 3.3 Å in both detergent micelles and lipid nanodiscs. CCB-bound GLUT4 exhibits an inward-open conformation. Despite the nearly identical conformation of the transmembrane domain to GLUT1, the cryo-EM structure reveals an extracellular glycosylation site and an intracellular helix that is invisible in the crystal structure of GLUT1. The structural study presented here lays the foundation for further mechanistic investigation of the modulation of GLUT4 trafficking. Our methods for cryo-EM analysis of GLUT4 will also facilitate structural determination of many other small size solute carriers. | |||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7wsm.cif.gz 7wsm.cif.gz | 94 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7wsm.ent.gz pdb7wsm.ent.gz | 67.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7wsm.json.gz 7wsm.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ws/7wsm https://data.pdbj.org/pub/pdb/validation_reports/ws/7wsm ftp://data.pdbj.org/pub/pdb/validation_reports/ws/7wsm ftp://data.pdbj.org/pub/pdb/validation_reports/ws/7wsm | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  32760MC  7wsnC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein | Mass: 56108.199 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: SLC2A4, GLUT4 / Production host: Homo sapiens (human) / Gene: SLC2A4, GLUT4 / Production host:  Homo sapiens (human) / References: UniProt: P14672 Homo sapiens (human) / References: UniProt: P14672 |
|---|---|
| #2: Polysaccharide | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose Source method: isolated from a genetically manipulated source |
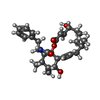
| #3: Chemical | ChemComp-5RH / |
| Has ligand of interest | Y |
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: GLUT4 / Type: COMPLEX / Entity ID: #1 / Source: RECOMBINANT |
|---|---|
| Molecular weight | Value: 56 kDa/nm / Experimental value: NO |
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Source (recombinant) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Buffer solution | pH: 8 |
| Specimen | Conc.: 1.8 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal defocus max: 2500 nm / Nominal defocus min: 800 nm |
| Image recording | Electron dose: 50 e/Å2 / Film or detector model: GATAN K3 (6k x 4k) |
- Processing
Processing
| Software | Name: PHENIX / Version: 1.18_3855: / Classification: refinement | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software |
| ||||||||||||||||||||||||
| CTF correction | Type: NONE | ||||||||||||||||||||||||
| Symmetry | Point symmetry: C1 (asymmetric) | ||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3.25 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 73000 / Symmetry type: POINT | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller



 PDBj
PDBj