+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7wfr | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human Nav1.8 with A-803467, class III | |||||||||||||||||||||||||||
 Components Components | Sodium channel protein type 10 subunit alpha | |||||||||||||||||||||||||||
 Keywords Keywords | MEMBRANE PROTEIN / Voltage-gated sodium channel / Nav / activation / selectivity | |||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationbundle of His cell action potential / AV node cell action potential / clathrin complex / regulation of atrial cardiac muscle cell membrane depolarization / membrane depolarization during action potential / voltage-gated monoatomic ion channel activity involved in regulation of presynaptic membrane potential / regulation of monoatomic ion transmembrane transport / sensory perception / cardiac muscle cell action potential involved in contraction / voltage-gated sodium channel complex ...bundle of His cell action potential / AV node cell action potential / clathrin complex / regulation of atrial cardiac muscle cell membrane depolarization / membrane depolarization during action potential / voltage-gated monoatomic ion channel activity involved in regulation of presynaptic membrane potential / regulation of monoatomic ion transmembrane transport / sensory perception / cardiac muscle cell action potential involved in contraction / voltage-gated sodium channel complex / Interaction between L1 and Ankyrins / voltage-gated sodium channel activity / odontogenesis of dentin-containing tooth / Phase 0 - rapid depolarisation / regulation of cardiac muscle contraction / regulation of heart rate / sodium ion transmembrane transport / presynaptic membrane / transmembrane transporter binding / axon / glutamatergic synapse / extracellular exosome / plasma membrane Similarity search - Function | |||||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3 Å | |||||||||||||||||||||||||||
 Authors Authors | Yan, N. / Pan, X.J. / Huang, X.S. / Huang, G.X. | |||||||||||||||||||||||||||
| Funding support |  China, 1items China, 1items
| |||||||||||||||||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2022 Journal: Proc Natl Acad Sci U S A / Year: 2022Title: Structural basis for high-voltage activation and subtype-specific inhibition of human Na1.8. Authors: Xiaoshuang Huang / Xueqin Jin / Gaoxingyu Huang / Jian Huang / Tong Wu / Zhangqiang Li / Jiaofeng Chen / Fang Kong / Xiaojing Pan / Nieng Yan /  Abstract: The dorsal root ganglia-localized voltage-gated sodium (Na) channel Na1.8 represents a promising target for developing next-generation analgesics. A prominent characteristic of Na1.8 is the ...The dorsal root ganglia-localized voltage-gated sodium (Na) channel Na1.8 represents a promising target for developing next-generation analgesics. A prominent characteristic of Na1.8 is the requirement of more depolarized membrane potential for activation. Here we present the cryogenic electron microscopy structures of human Na1.8 alone and bound to a selective pore blocker, A-803467, at overall resolutions of 2.7 to 3.2 Å. The first voltage-sensing domain (VSD) displays three different conformations. Structure-guided mutagenesis identified the extracellular interface between VSD and the pore domain (PD) to be a determinant for the high-voltage dependence of activation. A-803467 was clearly resolved in the central cavity of the PD, clenching S6. Our structure-guided functional characterizations show that two nonligand binding residues, Thr397 on S6 and Gly1406 on S6, allosterically modulate the channel's sensitivity to A-803467. Comparison of available structures of human Na channels suggests the extracellular loop region to be a potential site for developing subtype-specific pore-blocking biologics. | |||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7wfr.cif.gz 7wfr.cif.gz | 249.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7wfr.ent.gz pdb7wfr.ent.gz | 185.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7wfr.json.gz 7wfr.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/wf/7wfr https://data.pdbj.org/pub/pdb/validation_reports/wf/7wfr ftp://data.pdbj.org/pub/pdb/validation_reports/wf/7wfr ftp://data.pdbj.org/pub/pdb/validation_reports/wf/7wfr | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  32475MC  7we4C  7welC  7wfwC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 220903.500 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: SCN10A / Production host: Homo sapiens (human) / Gene: SCN10A / Production host:  Homo sapiens (human) / References: UniProt: Q9Y5Y9 Homo sapiens (human) / References: UniProt: Q9Y5Y9 |
|---|
-Sugars , 2 types, 5 molecules 
| #2: Polysaccharide | Source method: isolated from a genetically manipulated source #3: Sugar | |
|---|
-Non-polymers , 5 types, 26 molecules 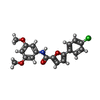








| #4: Chemical | ChemComp-95T / | ||||||
|---|---|---|---|---|---|---|---|
| #5: Chemical | ChemComp-CLR / #6: Chemical | ChemComp-PCW / #7: Chemical | ChemComp-LPE / #8: Chemical | ChemComp-P5S / | |
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: sodium channel III / Type: COMPLEX / Entity ID: #1 / Source: RECOMBINANT |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Source (recombinant) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Buffer solution | pH: 7.5 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal defocus max: 1800 nm / Nominal defocus min: 1500 nm |
| Image recording | Electron dose: 50 e/Å2 / Film or detector model: GATAN K3 (6k x 4k) |
- Processing
Processing
| Software | Name: PHENIX / Version: 1.18_3855: / Classification: refinement | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software | Name: PHENIX / Category: model refinement | ||||||||||||||||||||||||
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 274882 / Symmetry type: POINT | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller






 PDBj
PDBj















