+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human Nav1.8 with A-803467, class II | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Voltage-gated sodium channel / Nav / activation / selectivity / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationbundle of His cell action potential / AV node cell action potential / clathrin complex / regulation of atrial cardiac muscle cell membrane depolarization / membrane depolarization during action potential / regulation of monoatomic ion transmembrane transport / voltage-gated monoatomic ion channel activity involved in regulation of presynaptic membrane potential / sensory perception / cardiac muscle cell action potential involved in contraction / voltage-gated sodium channel complex ...bundle of His cell action potential / AV node cell action potential / clathrin complex / regulation of atrial cardiac muscle cell membrane depolarization / membrane depolarization during action potential / regulation of monoatomic ion transmembrane transport / voltage-gated monoatomic ion channel activity involved in regulation of presynaptic membrane potential / sensory perception / cardiac muscle cell action potential involved in contraction / voltage-gated sodium channel complex / Interaction between L1 and Ankyrins / voltage-gated sodium channel activity / odontogenesis of dentin-containing tooth / Phase 0 - rapid depolarisation / regulation of cardiac muscle contraction / regulation of heart rate / sodium ion transmembrane transport / presynaptic membrane / transmembrane transporter binding / axon / glutamatergic synapse / extracellular exosome / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Yan N / Pan XJ | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2022 Journal: Proc Natl Acad Sci U S A / Year: 2022Title: Structural basis for high-voltage activation and subtype-specific inhibition of human Na1.8. Authors: Xiaoshuang Huang / Xueqin Jin / Gaoxingyu Huang / Jian Huang / Tong Wu / Zhangqiang Li / Jiaofeng Chen / Fang Kong / Xiaojing Pan / Nieng Yan /  Abstract: The dorsal root ganglia-localized voltage-gated sodium (Na) channel Na1.8 represents a promising target for developing next-generation analgesics. A prominent characteristic of Na1.8 is the ...The dorsal root ganglia-localized voltage-gated sodium (Na) channel Na1.8 represents a promising target for developing next-generation analgesics. A prominent characteristic of Na1.8 is the requirement of more depolarized membrane potential for activation. Here we present the cryogenic electron microscopy structures of human Na1.8 alone and bound to a selective pore blocker, A-803467, at overall resolutions of 2.7 to 3.2 Å. The first voltage-sensing domain (VSD) displays three different conformations. Structure-guided mutagenesis identified the extracellular interface between VSD and the pore domain (PD) to be a determinant for the high-voltage dependence of activation. A-803467 was clearly resolved in the central cavity of the PD, clenching S6. Our structure-guided functional characterizations show that two nonligand binding residues, Thr397 on S6 and Gly1406 on S6, allosterically modulate the channel's sensitivity to A-803467. Comparison of available structures of human Na channels suggests the extracellular loop region to be a potential site for developing subtype-specific pore-blocking biologics. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32451.map.gz emd_32451.map.gz | 49.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32451-v30.xml emd-32451-v30.xml emd-32451.xml emd-32451.xml | 15.9 KB 15.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_32451.png emd_32451.png | 77.6 KB | ||
| Filedesc metadata |  emd-32451.cif.gz emd-32451.cif.gz | 7.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32451 http://ftp.pdbj.org/pub/emdb/structures/EMD-32451 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32451 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32451 | HTTPS FTP |
-Validation report
| Summary document |  emd_32451_validation.pdf.gz emd_32451_validation.pdf.gz | 413.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_32451_full_validation.pdf.gz emd_32451_full_validation.pdf.gz | 412.6 KB | Display | |
| Data in XML |  emd_32451_validation.xml.gz emd_32451_validation.xml.gz | 6 KB | Display | |
| Data in CIF |  emd_32451_validation.cif.gz emd_32451_validation.cif.gz | 6.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32451 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32451 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32451 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32451 | HTTPS FTP |
-Related structure data
| Related structure data |  7welMC  7we4C  7wfrC  7wfwC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32451.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32451.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0825 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : sodium channel II
| Entire | Name: sodium channel II |
|---|---|
| Components |
|
-Supramolecule #1: sodium channel II
| Supramolecule | Name: sodium channel II / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Sodium channel protein type 10 subunit alpha
| Macromolecule | Name: Sodium channel protein type 10 subunit alpha / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 220.9035 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEFPIGSLET NNFRRFTPES LVEIEKQIAA KQGTKKAREK HREQKDQEEK PRPQLDLKAC NQLPKFYGEL PAELIGEPLE DLDPFYSTH RTFMVLNKGR TISRFSATRA LWLFSPFNLI RRTAIKVSVH SWFSLFITVT ILVNCVCMTR TDLPEKIEYV F TVIYTFEA ...String: MEFPIGSLET NNFRRFTPES LVEIEKQIAA KQGTKKAREK HREQKDQEEK PRPQLDLKAC NQLPKFYGEL PAELIGEPLE DLDPFYSTH RTFMVLNKGR TISRFSATRA LWLFSPFNLI RRTAIKVSVH SWFSLFITVT ILVNCVCMTR TDLPEKIEYV F TVIYTFEA LIKILARGFC LNEFTYLRDP WNWLDFSVIT LAYVGTAIDL RGISGLRTFR VLRALKTVSV IPGLKVIVGA LI HSVKKLA DVTILTIFCL SVFALVGLQL FKGNLKNKCV KNDMAVNETT NYSSHRKPDI YINKRGTSDP LLCGNGSDSG HCP DGYICL KTSDNPDFNY TSFDSFAWAF LSLFRLMTQD SWERLYQQTL RTSGKIYMIF FVLVIFLGSF YLVNLILAVV TMAY EEQNQ ATTDEIEAKE KKFQEALEML RKEQEVLAAL GIDTTSLHSH NGSPLTSKNA SERRHRIKPR VSEGSTEDNK SPRSD PYNQ RRMSFLGLAS GKRRASHGSV FHFRSPGRDI SLPEGVTDDG VFPGDHESHR GSLLLGGGAG QQGPLPRSPL PQPSNP DSR HGEDEHQPPP TSELAPGAVD VSAFDAGQKK TFLSAEYLDE PFRAQRAMSV VSIITSVLEE LEESEQKCPP CLTSLSQ KY LIWDCCPMWV KLKTILFGLV TDPFAELTIT LCIVVNTIFM AMEHHGMSPT FEAMLQIGNI VFTIFFTAEM VFKIIAFD P YYYFQKKWNI FDCIIVTVSL LELGVAKKGS LSVLRSFRLL RVFKLAKSWP TLNTLIKIIG NSVGALGNLT IILAIIVFV FALVGKQLLG ENYRNNRKNI SAPHEDWPRW HMHDFFHSFL IVFRILCGEW IENMWACMEV GQKSICLILF LTVMVLGNLV VLNLFIALL LNSFFADNLT APEDDGEVNN LQVALARIQV FGHRTKQALC SFFSRSCPFP QPKAEPELVV KLPLSSSKAE N HIAANTAR GSSGGLQAPR GPRDEHSDFI ANPTVWVSVP IAEGESDLDD LEDDGGEDAQ SFQQEVIPKG QQEQLQQVER CG DHLTPRS PGTGTSSEDL APSLGETWKD ESVPQVPAEG VDDTSSSEGS TVDCLDPEEI LRKIPELADD LEEPDDCFTE GCI RHCPCC KLDTTKSPWD VGWQVRKTCY RIVEHSWFES FIIFMILLSS GSLAFEDYYL DQKPTVKALL EYTDRVFTFI FVFE MLLKW VAYGFKKYFT NAWCWLDFLI VNISLISLTA KILEYSEVAP IKALRTLRAL RPLRALSRFE GMRVVVDALV GAIPS IMNV LLVCLIFWLI FSIMGVNLFA GKFWRCINYT DGEFSLVPLS IVNNKSDCKI QNSTGSFFWV NVKVNFDNVA MGYLAL LQV ATFKGWMDIM YAAVDSREVN MQPKWEDNVY MYLYFVIFII FGGFFTLNLF VGVIIDNFNQ QKKKLGGQDI FMTEEQK KY YNAMKKLGSK KPQKPIPRPL NKFQGFVFDI VTRQAFDITI MVLICLNMIT MMVETDDQSE EKTKILGKIN QFFVAVFT G ECVMKMFALR QYYFTNGWNV FDFIVVVLSI ASLIFSAILK SLQSYFSPTL FRVIRLARIG RILRLIRAAK GIRTLLFAL MMSLPALFNI GLLLFLVMFI YSIFGMSSFP HVRWEAGIDD MFNFQTFANS MLCLFQITTS AGWDGLLSPI LNTGPPYCDP NLPNSNGTR GDCGSPAVGI IFFTTYIIIS FLIMVNMYIA VILENFNVAT EESTEPLSED DFDMFYETWE KFDPEATQFI T FSALSDFA DTLSGPLRIP KPNRNILIQM DLPLVPGDKI HCLDILFAFT KNVLGESGEL DSLKANMEEK FMATNLSKSS YE PIATTLR WKQEDISATV IQKAYRSYVL HRSMALSNTP CVPRAEEEAA SLPDEGFVAF TANENCVLPD KSETASATSF PPS YESVTR GLSDRVNMRT SSSIQNEDEA TSMELIAPGP UniProtKB: Sodium channel protein type 10 subunit alpha |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 3 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #4: 5-(4-chlorophenyl)-~{N}-(3,5-dimethoxyphenyl)furan-2-carboxamide
| Macromolecule | Name: 5-(4-chlorophenyl)-~{N}-(3,5-dimethoxyphenyl)furan-2-carboxamide type: ligand / ID: 4 / Number of copies: 1 / Formula: 95T |
|---|---|
| Molecular weight | Theoretical: 357.788 Da |
| Chemical component information | 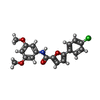 ChemComp-95T: |
-Macromolecule #5: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 5 / Number of copies: 5 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Macromolecule #6: 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE
| Macromolecule | Name: 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE / type: ligand / ID: 6 / Number of copies: 7 / Formula: PCW |
|---|---|
| Molecular weight | Theoretical: 787.121 Da |
| Chemical component information |  ChemComp-PCW: |
-Macromolecule #7: 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE
| Macromolecule | Name: 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE / type: ligand / ID: 7 / Number of copies: 12 / Formula: LPE |
|---|---|
| Molecular weight | Theoretical: 510.708 Da |
| Chemical component information |  ChemComp-LPE: |
-Macromolecule #8: O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phospho...
| Macromolecule | Name: O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine type: ligand / ID: 8 / Number of copies: 2 / Formula: P5S |
|---|---|
| Molecular weight | Theoretical: 792.075 Da |
| Chemical component information |  ChemComp-P5S: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




















