+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7w4o | ||||||
|---|---|---|---|---|---|---|---|
| Title | The structure of KATP H175K mutant in pre-open state | ||||||
 Components Components |
| ||||||
 Keywords Keywords | TRANSPORT PROTEIN / KATP / Kir6.2 / SUR1 / NN414 / ADP / ATP | ||||||
| Function / homology |  Function and homology information Function and homology informationATP sensitive Potassium channels / ATP-activated inward rectifier potassium channel activity / glutamate secretion, neurotransmission / response to resveratrol / inward rectifying potassium channel / Regulation of insulin secretion / sulfonylurea receptor activity / ventricular cardiac muscle tissue development / cell body fiber / ABC-family proteins mediated transport ...ATP sensitive Potassium channels / ATP-activated inward rectifier potassium channel activity / glutamate secretion, neurotransmission / response to resveratrol / inward rectifying potassium channel / Regulation of insulin secretion / sulfonylurea receptor activity / ventricular cardiac muscle tissue development / cell body fiber / ABC-family proteins mediated transport / CAMKK-AMPK signaling cascade / voltage-gated monoatomic ion channel activity involved in regulation of presynaptic membrane potential / ATPase-coupled monoatomic cation transmembrane transporter activity / inward rectifier potassium channel activity / Ion homeostasis / nervous system process / : / ankyrin binding / neuromuscular process / response to testosterone / response to ATP / potassium ion import across plasma membrane / potassium ion binding / response to stress / intercalated disc / action potential / axolemma / potassium channel activity / ABC-type transporter activity / positive regulation of insulin secretion involved in cellular response to glucose stimulus / cellular response to nutrient levels / heat shock protein binding / T-tubule / acrosomal vesicle / response to ischemia / determination of adult lifespan / positive regulation of protein localization to plasma membrane / cellular response to glucose stimulus / negative regulation of insulin secretion / ADP binding / sarcolemma / cellular response to nicotine / glucose metabolic process / cellular response to tumor necrosis factor / nuclear envelope / response to estradiol / presynapse / presynaptic membrane / transmembrane transporter binding / response to hypoxia / endosome / response to xenobiotic stimulus / neuronal cell body / apoptotic process / glutamatergic synapse / ATP hydrolysis activity / ATP binding / metal ion binding / plasma membrane / cytoplasm Similarity search - Function | ||||||
| Biological species |   Mesocricetus auratus (golden hamster) Mesocricetus auratus (golden hamster) | ||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 2.96 Å | ||||||
 Authors Authors | Chen, L. / Wang, M. | ||||||
| Funding support |  China, 1items China, 1items
| ||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structural insights into the mechanism of pancreatic K channel regulation by nucleotides. Authors: Mengmeng Wang / Jing-Xiang Wu / Dian Ding / Lei Chen /  Abstract: ATP-sensitive potassium channels (K) are metabolic sensors that convert the intracellular ATP/ADP ratio to the excitability of cells. They are involved in many physiological processes and implicated ...ATP-sensitive potassium channels (K) are metabolic sensors that convert the intracellular ATP/ADP ratio to the excitability of cells. They are involved in many physiological processes and implicated in several human diseases. Here we present the cryo-EM structures of the pancreatic K channel in both the closed state and the pre-open state, resolved in the same sample. We observe the binding of nucleotides at the inhibitory sites of the Kir6.2 channel in the closed but not in the pre-open state. Structural comparisons reveal the mechanism for ATP inhibition and Mg-ADP activation, two fundamental properties of K channels. Moreover, the structures also uncover the activation mechanism of diazoxide-type K openers. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7w4o.cif.gz 7w4o.cif.gz | 1.3 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7w4o.ent.gz pdb7w4o.ent.gz | 1.1 MB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7w4o.json.gz 7w4o.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/w4/7w4o https://data.pdbj.org/pub/pdb/validation_reports/w4/7w4o ftp://data.pdbj.org/pub/pdb/validation_reports/w4/7w4o ftp://data.pdbj.org/pub/pdb/validation_reports/w4/7w4o | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  32310MC  7w4pC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Protein , 2 types, 8 molecules ACEGBDFH
| #1: Protein | Mass: 43606.770 Da / Num. of mol.: 4 / Mutation: H175K Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / References: UniProt: Q61743 Homo sapiens (human) / References: UniProt: Q61743#2: Protein | Mass: 177295.516 Da / Num. of mol.: 4 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Mesocricetus auratus (golden hamster) / Gene: Abcc8 / Production host: Mesocricetus auratus (golden hamster) / Gene: Abcc8 / Production host:  Homo sapiens (human) / References: UniProt: A0A1U7R319 Homo sapiens (human) / References: UniProt: A0A1U7R319 |
|---|
-Non-polymers , 4 types, 20 molecules 

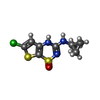




| #3: Chemical | ChemComp-ADP / #4: Chemical | ChemComp-MG / #5: Chemical | ChemComp-E2H / #6: Chemical | ChemComp-ATP / |
|---|
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Source (natural) |
| ||||||||||||||||||||||||
| Source (recombinant) |
| ||||||||||||||||||||||||
| Buffer solution | pH: 7.5 | ||||||||||||||||||||||||
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES | ||||||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal defocus max: 2500 nm / Nominal defocus min: 1000 nm |
| Image recording | Electron dose: 50 e/Å2 / Film or detector model: GATAN K2 SUMMIT (4k x 4k) |
- Processing
Processing
| CTF correction | Type: NONE |
|---|---|
| 3D reconstruction | Resolution: 2.96 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 216000 Details: This is a composite map. Map resolutions are "2.87/2.96/2.72" for each individual map. Symmetry type: POINT |
 Movie
Movie Controller
Controller




 PDBj
PDBj










