[English] 日本語
 Yorodumi
Yorodumi- PDB-7vzg: Structure of the Acidobacteria homodimeric reaction center bound ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7vzg | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the Acidobacteria homodimeric reaction center bound with cytochrome c (the larger form) | |||||||||||||||
 Components Components |
| |||||||||||||||
 Keywords Keywords | PHOTOSYNTHESIS / membrane protein | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationthylakoid / iron-sulfur cluster binding / photosynthesis / electron transfer activity / iron ion binding / heme binding / metal ion binding / membrane Similarity search - Function | |||||||||||||||
| Biological species |  Chloracidobacterium thermophilum (bacteria) Chloracidobacterium thermophilum (bacteria) | |||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 2.61 Å | |||||||||||||||
 Authors Authors | Huang, G.Q. / Dong, S.S. / Qin, X.C. / Sui, S.F. | |||||||||||||||
| Funding support | 1items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structure of the Acidobacteria homodimeric reaction center bound with cytochrome c. Authors: Shishang Dong / Guoqiang Huang / Changhui Wang / Jiajia Wang / Sen-Fang Sui / Xiaochun Qin /  Abstract: Photosynthesis converts light energy to chemical energy to fuel life on earth. Light energy is harvested by antenna pigments and transferred to reaction centers (RCs) to drive the electron transfer ...Photosynthesis converts light energy to chemical energy to fuel life on earth. Light energy is harvested by antenna pigments and transferred to reaction centers (RCs) to drive the electron transfer (ET) reactions. Here, we present cryo-electron microscopy (cryo-EM) structures of two forms of the RC from the microaerophilic Chloracidobacterium thermophilum (CabRC): one containing 10 subunits, including two different cytochromes; and the other possessing two additional subunits, PscB and PscZ. The larger form contained 2 Zn-bacteriochlorophylls, 16 bacteriochlorophylls, 10 chlorophylls, 2 lycopenes, 2 hemes, 3 FeS clusters, 12 lipids, 2 Ca ions and 6 water molecules, revealing a type I RC with an ET chain involving two hemes and a hybrid antenna containing bacteriochlorophylls and chlorophylls. Our results provide a structural basis for understanding the excitation energy and ET within the CabRC and offer evolutionary insights into the origin and adaptation of photosynthetic RCs. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7vzg.cif.gz 7vzg.cif.gz | 513.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7vzg.ent.gz pdb7vzg.ent.gz | 401.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7vzg.json.gz 7vzg.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/vz/7vzg https://data.pdbj.org/pub/pdb/validation_reports/vz/7vzg ftp://data.pdbj.org/pub/pdb/validation_reports/vz/7vzg ftp://data.pdbj.org/pub/pdb/validation_reports/vz/7vzg | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  32228MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Protein , 5 types, 6 molecules AaCcBD
| #1: Protein | Mass: 98537.555 Da / Num. of mol.: 2 / Source method: isolated from a natural source Source: (natural)  Chloracidobacterium thermophilum (bacteria) Chloracidobacterium thermophilum (bacteria)References: UniProt: G2LDR8 #2: Protein | | Mass: 22369.820 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  Chloracidobacterium thermophilum (bacteria) Chloracidobacterium thermophilum (bacteria)References: UniProt: G2LDR4 #7: Protein | | Mass: 15664.669 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  Chloracidobacterium thermophilum (bacteria) Chloracidobacterium thermophilum (bacteria)References: UniProt: G2LDR3 #8: Protein | | Mass: 8199.330 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  Chloracidobacterium thermophilum (bacteria) Chloracidobacterium thermophilum (bacteria)References: UniProt: A8DJF8 #9: Protein | | Mass: 8622.735 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  Chloracidobacterium thermophilum (bacteria) Chloracidobacterium thermophilum (bacteria)References: UniProt: G2LHG2 |
|---|
-Protein/peptide , 4 types, 8 molecules EeFfGgHh
| #3: Protein/peptide | Mass: 3684.458 Da / Num. of mol.: 2 / Source method: isolated from a natural source Source: (natural)  Chloracidobacterium thermophilum (bacteria) Chloracidobacterium thermophilum (bacteria)References: UniProt: G2LK98 #4: Protein/peptide | Mass: 3885.684 Da / Num. of mol.: 2 / Source method: isolated from a natural source Source: (natural)  Chloracidobacterium thermophilum (bacteria) Chloracidobacterium thermophilum (bacteria)References: UniProt: G2LEN5 #5: Protein/peptide | Mass: 4293.273 Da / Num. of mol.: 2 / Source method: isolated from a natural source Source: (natural)  Chloracidobacterium thermophilum (bacteria) Chloracidobacterium thermophilum (bacteria)References: UniProt: G2LJ20 #6: Protein/peptide | Mass: 1635.006 Da / Num. of mol.: 2 / Source method: isolated from a natural source Source: (natural)  Chloracidobacterium thermophilum (bacteria) Chloracidobacterium thermophilum (bacteria) |
|---|
-Non-polymers , 12 types, 87 molecules 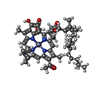



















| #10: Chemical | | #11: Chemical | ChemComp-BCL / #12: Chemical | ChemComp-CLA / #13: Chemical | #14: Chemical | #15: Chemical | ChemComp-85I / [( #16: Chemical | ChemComp-UNL / Mass: 663.906 Da / Num. of mol.: 32 / Source method: obtained synthetically / Feature type: SUBJECT OF INVESTIGATION #17: Chemical | ChemComp-85N / [( #18: Chemical | #19: Chemical | #20: Chemical | #21: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: CabRC complex, the larger form / Type: COMPLEX / Entity ID: #1-#9 / Source: NATURAL |
|---|---|
| Source (natural) | Organism:  Chloracidobacterium thermophilum (bacteria) Chloracidobacterium thermophilum (bacteria) |
| Buffer solution | pH: 7.6 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal defocus max: 2000 nm / Nominal defocus min: 500 nm / Cs: 2.7 mm |
| Image recording | Electron dose: 50 e/Å2 / Film or detector model: GATAN K3 (6k x 4k) |
- Processing
Processing
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION |
|---|---|
| 3D reconstruction | Resolution: 2.61 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 52612 / Symmetry type: POINT |
 Movie
Movie Controller
Controller


 PDBj
PDBj




























