+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
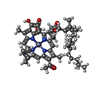
| タイトル | Structure of the Acidobacteria homodimeric reaction center bound with cytochrome c (the larger form) | |||||||||
 マップデータ マップデータ | ||||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | membrane protein / PHOTOSYNTHESIS | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報thylakoid / iron-sulfur cluster binding / photosynthesis / electron transfer activity / iron ion binding / heme binding / metal ion binding / membrane 類似検索 - 分子機能 | |||||||||
| 生物種 |  Chloracidobacterium thermophilum (バクテリア) Chloracidobacterium thermophilum (バクテリア) | |||||||||
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 2.61 Å | |||||||||
 データ登録者 データ登録者 | Huang GQ / Dong SS / Qin XC / Sui SF | |||||||||
| 資金援助 | 1件
| |||||||||
 引用 引用 |  ジャーナル: Nat Commun / 年: 2022 ジャーナル: Nat Commun / 年: 2022タイトル: Structure of the Acidobacteria homodimeric reaction center bound with cytochrome c. 著者: Shishang Dong / Guoqiang Huang / Changhui Wang / Jiajia Wang / Sen-Fang Sui / Xiaochun Qin /  要旨: Photosynthesis converts light energy to chemical energy to fuel life on earth. Light energy is harvested by antenna pigments and transferred to reaction centers (RCs) to drive the electron transfer ...Photosynthesis converts light energy to chemical energy to fuel life on earth. Light energy is harvested by antenna pigments and transferred to reaction centers (RCs) to drive the electron transfer (ET) reactions. Here, we present cryo-electron microscopy (cryo-EM) structures of two forms of the RC from the microaerophilic Chloracidobacterium thermophilum (CabRC): one containing 10 subunits, including two different cytochromes; and the other possessing two additional subunits, PscB and PscZ. The larger form contained 2 Zn-bacteriochlorophylls, 16 bacteriochlorophylls, 10 chlorophylls, 2 lycopenes, 2 hemes, 3 FeS clusters, 12 lipids, 2 Ca ions and 6 water molecules, revealing a type I RC with an ET chain involving two hemes and a hybrid antenna containing bacteriochlorophylls and chlorophylls. Our results provide a structural basis for understanding the excitation energy and ET within the CabRC and offer evolutionary insights into the origin and adaptation of photosynthetic RCs. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_32228.map.gz emd_32228.map.gz | 38.4 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-32228-v30.xml emd-32228-v30.xml emd-32228.xml emd-32228.xml | 24.1 KB 24.1 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| 画像 |  emd_32228.png emd_32228.png | 78.5 KB | ||
| Filedesc metadata |  emd-32228.cif.gz emd-32228.cif.gz | 7.4 KB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32228 http://ftp.pdbj.org/pub/emdb/structures/EMD-32228 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32228 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32228 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_32228_validation.pdf.gz emd_32228_validation.pdf.gz | 545.3 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_32228_full_validation.pdf.gz emd_32228_full_validation.pdf.gz | 544.8 KB | 表示 | |
| XML形式データ |  emd_32228_validation.xml.gz emd_32228_validation.xml.gz | 6 KB | 表示 | |
| CIF形式データ |  emd_32228_validation.cif.gz emd_32228_validation.cif.gz | 6.9 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32228 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32228 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32228 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32228 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  7vzgMC M: このマップから作成された原子モデル C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_32228.map.gz / 形式: CCP4 / 大きさ: 40.6 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_32228.map.gz / 形式: CCP4 / 大きさ: 40.6 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.0825 Å | ||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
- 試料の構成要素
試料の構成要素
+全体 : CabRC complex, the larger form
+超分子 #1: CabRC complex, the larger form
+分子 #1: PscA
+分子 #2: Cytochrome c, mono-and diheme variants
+分子 #3: PscE
+分子 #4: PscF
+分子 #5: PscG
+分子 #6: undefined polypeptide
+分子 #7: Cytochrome c domain-containing protein
+分子 #8: Photosystem P840 reaction center iron-sulfur protein
+分子 #9: PscD'
+分子 #10: [methyl 9-acetyl-14-ethyl-20-hydroxy-4,8,13,18-tetramethyl-3-{3-o...
+分子 #11: BACTERIOCHLOROPHYLL A
+分子 #12: CHLOROPHYLL A
+分子 #13: LYCOPENE
+分子 #14: CALCIUM ION
+分子 #15: [(2~{R})-2-[2-(methylamino)ethoxy-oxidanyl-phosphoryl]oxy-2-(13-m...
+分子 #16: UNKNOWN LIGAND
+分子 #17: [(2~{S})-2-[[(1~{R})-1,2-bis(13-methyltetradecanoyloxy)ethoxy]met...
+分子 #18: HEME C
+分子 #19: [(2S)-2-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-2-(13-methyltetra...
+分子 #20: IRON/SULFUR CLUSTER
+分子 #21: water
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 7.6 |
|---|---|
| 凍結 | 凍結剤: ETHANE |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K3 (6k x 4k) / 平均電子線量: 50.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / Cs: 2.7 mm / 最大 デフォーカス(公称値): 2.0 µm / 最小 デフォーカス(公称値): 0.5 µm |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)