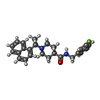
Entry Database : PDB / ID : 7tzjTitle SARS CoV-2 PLpro in complex with inhibitor 3k Papain-like protease Keywords / / / / / / / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Method / / / Resolution : 2.66 Å Authors Calleja, D.J. / Klemm, T. / Lechtenberg, B.C. / Kuchel, N.W. / Lessene, G. / Komander, D. Funding support Organization Grant number Country National Health and Medical Research Council (NHMRC, Australia) GNT1178122 Wellcome Trust WT222698/Z/21/Z National Health and Medical Research Council (NHMRC, Australia) MRF2002119 National Health and Medical Research Council (NHMRC, Australia) GNT1117089
Journal : Front Chem / Year : 2022Title : Insights Into Drug Repurposing, as Well as Specificity and Compound Properties of Piperidine-Based SARS-CoV-2 PLpro Inhibitors.Authors: Calleja, D.J. / Kuchel, N. / Lu, B.G.C. / Birkinshaw, R.W. / Klemm, T. / Doerflinger, M. / Cooney, J.P. / Mackiewicz, L. / Au, A.E. / Yap, Y.Q. / Blackmore, T.R. / Katneni, K. / Crighton, E. ... Authors : Calleja, D.J. / Kuchel, N. / Lu, B.G.C. / Birkinshaw, R.W. / Klemm, T. / Doerflinger, M. / Cooney, J.P. / Mackiewicz, L. / Au, A.E. / Yap, Y.Q. / Blackmore, T.R. / Katneni, K. / Crighton, E. / Newman, J. / Jarman, K.E. / Call, M.J. / Lechtenberg, B.C. / Czabotar, P.E. / Pellegrini, M. / Charman, S.A. / Lowes, K.N. / Mitchell, J.P. / Nachbur, U. / Lessene, G. / Komander, D. History Deposition Feb 15, 2022 Deposition site / Processing site Revision 1.0 Mar 2, 2022 Provider / Type Revision 1.1 May 11, 2022 Group / Category / citation_authorItem _citation.country / _citation.journal_abbrev ... _citation.country / _citation.journal_abbrev / _citation.journal_id_CSD / _citation.journal_id_ISSN / _citation.journal_volume / _citation.page_first / _citation.page_last / _citation.pdbx_database_id_DOI / _citation.pdbx_database_id_PubMed / _citation.title / _citation.year Revision 1.2 Oct 18, 2023 Group / Refinement descriptionCategory / chem_comp_bond / pdbx_initial_refinement_model
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.66 Å
MOLECULAR REPLACEMENT / Resolution: 2.66 Å  Authors
Authors Australia,
Australia,  United Kingdom, 4items
United Kingdom, 4items  Citation
Citation Journal: Front Chem / Year: 2022
Journal: Front Chem / Year: 2022 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 7tzj.cif.gz
7tzj.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb7tzj.ent.gz
pdb7tzj.ent.gz PDB format
PDB format 7tzj.json.gz
7tzj.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/tz/7tzj
https://data.pdbj.org/pub/pdb/validation_reports/tz/7tzj ftp://data.pdbj.org/pub/pdb/validation_reports/tz/7tzj
ftp://data.pdbj.org/pub/pdb/validation_reports/tz/7tzj
 Links
Links Assembly
Assembly


 Components
Components

 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  Australian Synchrotron
Australian Synchrotron  / Beamline: MX2 / Wavelength: 0.9537 Å
/ Beamline: MX2 / Wavelength: 0.9537 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller












 PDBj
PDBj