[English] 日本語
 Yorodumi
Yorodumi- PDB-7r3k: Chlamydomonas reinhardtii TSP9 mutant small Photosystem I complex -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7r3k | ||||||
|---|---|---|---|---|---|---|---|
| Title | Chlamydomonas reinhardtii TSP9 mutant small Photosystem I complex | ||||||
 Components Components |
| ||||||
 Keywords Keywords | PHOTOSYNTHESIS / Photosystem I / Complex / Green algae | ||||||
| Function / homology |  Function and homology information Function and homology informationphotosynthesis, light harvesting in photosystem I / photosynthesis, light harvesting / chloroplast thylakoid lumen / photosystem I reaction center / photosystem I / photosynthetic electron transport in photosystem I / photosystem I / photosystem II / chlorophyll binding / chloroplast thylakoid membrane ...photosynthesis, light harvesting in photosystem I / photosynthesis, light harvesting / chloroplast thylakoid lumen / photosystem I reaction center / photosystem I / photosynthetic electron transport in photosystem I / photosystem I / photosystem II / chlorophyll binding / chloroplast thylakoid membrane / response to light stimulus / photosynthesis / 4 iron, 4 sulfur cluster binding / electron transfer activity / oxidoreductase activity / magnesium ion binding / metal ion binding Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 2.52 Å | ||||||
 Authors Authors | Klaiman, D. / Schwartz, T. / Nelson, N. | ||||||
| Funding support |  Israel, 1items Israel, 1items
| ||||||
 Citation Citation |  Journal: Biomolecules / Year: 2023 Journal: Biomolecules / Year: 2023Title: Structure of Photosystem I Supercomplex Isolated from a Cytochrome b6f Temperature-Sensitive Mutant. Authors: Tom Schwartz / Mariia Fadeeva / Daniel Klaiman / Nathan Nelson /  Abstract: The unicellular green alga, , has been widely used as a model system to study photosynthesis. Its possibility to generate and analyze specific mutants has made it an excellent tool for mechanistic ...The unicellular green alga, , has been widely used as a model system to study photosynthesis. Its possibility to generate and analyze specific mutants has made it an excellent tool for mechanistic and biogenesis studies. Using negative selection of ultraviolet (UV) irradiation-mutated cells, we isolated a mutant (TSP9) with a single amino acid mutation in the Rieske protein of the cytochrome b6f complex. The W143R mutation in the petC gene resulted in total loss of cytochrome b6f complex function at the non-permissive temperature of 37 °C and recovery at the permissive temperature of 25 °C. We then isolated photosystem I (PSI) and photosystem II (PSII) supercomplexes from cells grown at the non-permissive temperature and determined the PSI structure with high-resolution cryogenic electron microscopy. There were several structural alterations compared with the structures obtained from wild-type cells. Our structural data suggest that the mutant responded by excluding the Lhca2, Lhca9, PsaL, and PsaH subunits. This structural alteration prevents state two transition, where LHCII migrates from PSII to bind to the PSI complex. We propose this as a possible response mechanism triggered by the TSP9 phenotype at the non-permissive temperature. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7r3k.cif.gz 7r3k.cif.gz | 1 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7r3k.ent.gz pdb7r3k.ent.gz | 931.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7r3k.json.gz 7r3k.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/r3/7r3k https://data.pdbj.org/pub/pdb/validation_reports/r3/7r3k ftp://data.pdbj.org/pub/pdb/validation_reports/r3/7r3k ftp://data.pdbj.org/pub/pdb/validation_reports/r3/7r3k | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  14248MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Photosystem I P700 chlorophyll a apoprotein ... , 2 types, 2 molecules AB
| #1: Protein | Mass: 83239.203 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|---|
| #2: Protein | Mass: 82184.266 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
-Protein , 1 types, 1 molecules C
| #3: Protein | Mass: 8869.325 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|
-Photosystem I reaction center subunit ... , 7 types, 7 molecules DEFGIJK
| #4: Protein | Mass: 21401.885 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|---|
| #5: Protein | Mass: 10786.395 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #6: Protein | Mass: 24088.936 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #7: Protein | Mass: 13236.007 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #8: Protein | Mass: 10586.388 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #9: Protein/peptide | Mass: 4750.509 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #10: Protein | Mass: 11214.084 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
-Chlorophyll a-b binding protein, ... , 7 types, 8 molecules 1Z378456
| #11: Protein | Mass: 23923.205 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  #12: Protein | | Mass: 32629.486 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  #13: Protein | | Mass: 26248.869 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  #14: Protein | | Mass: 25948.812 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  #15: Protein | | Mass: 28729.822 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  #16: Protein | | Mass: 28257.555 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  #17: Protein | | Mass: 27812.373 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|
-Sugars , 2 types, 17 molecules 


| #24: Sugar | ChemComp-LMT / #25: Sugar | ChemComp-DGD / | |
|---|
-Non-polymers , 20 types, 867 molecules 







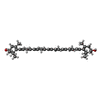






























| #18: Chemical | ChemComp-CL0 / | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| #19: Chemical | ChemComp-CLA / #20: Chemical | #21: Chemical | #22: Chemical | ChemComp-BCR / #23: Chemical | ChemComp-LHG / #26: Chemical | ChemComp-CA / | #27: Chemical | #28: Chemical | #29: Chemical | #30: Chemical | ChemComp-DAO / | #31: Chemical | ChemComp-LUT / ( #32: Chemical | ChemComp-CHL / #33: Chemical | #34: Chemical | ChemComp-3PH / #35: Chemical | ChemComp-DGA / | #36: Chemical | ChemComp-PLM / | #37: Chemical | ChemComp-SQD / | #38: Chemical | #39: Water | ChemComp-HOH / | |
-Details
| Has ligand of interest | N |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: C. reinhardtii Photosystem I supercomplex / Type: COMPLEX / Details: Contains 10 core subunits and 8 antenna proteins / Entity ID: #1-#17 / Source: NATURAL |
|---|---|
| Molecular weight | Value: 0.78 MDa / Experimental value: YES |
| Source (natural) | Organism:  |
| Buffer solution | pH: 8 |
| Specimen | Conc.: 3 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES / Details: Chlorophyll concentration was provided |
| Specimen support | Details: Harrick Plasma cleaner PDC-32G-2 / Grid material: COPPER / Grid mesh size: 300 divisions/in. / Grid type: Quantifoil R1.2/1.3 |
| Vitrification | Instrument: LEICA EM GP / Cryogen name: ETHANE / Humidity: 90 % / Chamber temperature: 295 K / Details: blot for 3 seconds before plunging |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 105000 X / Nominal defocus max: 9000 nm / Nominal defocus min: 500 nm / Cs: 2.7 mm / C2 aperture diameter: 50 µm |
| Image recording | Average exposure time: 2.6 sec. / Electron dose: 57.74 e/Å2 / Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Num. of real images: 13173 |
- Processing
Processing
| Software | Name: PHENIX / Version: 1.19.2_4158: / Classification: refinement | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software |
| ||||||||||||||||||||||||||||||||||||
| CTF correction | Type: NONE | ||||||||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 1009378 | ||||||||||||||||||||||||||||||||||||
| Symmetry | Point symmetry: C1 (asymmetric) | ||||||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 2.52 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 173187 / Num. of class averages: 3 / Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||
| Atomic model building | Protocol: RIGID BODY FIT / Space: REAL | ||||||||||||||||||||||||||||||||||||
| Atomic model building | PDB-ID: 6JO5 Accession code: 6JO5 / Source name: PDB / Type: experimental model | ||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller


 PDBj
PDBj







































