[English] 日本語
 Yorodumi
Yorodumi- PDB-7prv: The glucocorticoid receptor in complex with fluticasone furoate, ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7prv | ||||||
|---|---|---|---|---|---|---|---|
| Title | The glucocorticoid receptor in complex with fluticasone furoate, a PGC1a coactivator fragment and sgk 23bp | ||||||
 Components Components |
| ||||||
 Keywords Keywords | SIGNALING PROTEIN / nuclear receptor / ligand-activated transcription factor / multi-domain / complex / agonist | ||||||
| Function / homology |  Function and homology information Function and homology informationRegulation of MITF-M dependent genes involved in metabolism / Regulation of NPAS4 gene transcription / regulation of glucocorticoid biosynthetic process / nuclear glucocorticoid receptor activity / steroid hormone binding / glucocorticoid metabolic process / response to cortisol / PTK6 Expression / neuroinflammatory response / positive regulation of fatty acid oxidation ...Regulation of MITF-M dependent genes involved in metabolism / Regulation of NPAS4 gene transcription / regulation of glucocorticoid biosynthetic process / nuclear glucocorticoid receptor activity / steroid hormone binding / glucocorticoid metabolic process / response to cortisol / PTK6 Expression / neuroinflammatory response / positive regulation of fatty acid oxidation / mammary gland duct morphogenesis / microglia differentiation / cellular respiration / maternal behavior / astrocyte differentiation / Activation of PPARGC1A (PGC-1alpha) by phosphorylation / adrenal gland development / regulation of gluconeogenesis / cellular response to glucocorticoid stimulus / cellular response to steroid hormone stimulus / response to starvation / temperature homeostasis / response to muscle activity / lncRNA binding / intracellular glucose homeostasis / fatty acid oxidation / response to dietary excess / motor behavior / estrogen response element binding / cellular response to dexamethasone stimulus / adipose tissue development / nuclear receptor-mediated steroid hormone signaling pathway / FOXO-mediated transcription of oxidative stress, metabolic and neuronal genes / cellular response to transforming growth factor beta stimulus / brown fat cell differentiation / core promoter sequence-specific DNA binding / Transcriptional regulation of brown and beige adipocyte differentiation by EBF2 / energy homeostasis / digestion / steroid binding / positive regulation of gluconeogenesis / SUMOylation of transcription cofactors / HSP90 chaperone cycle for steroid hormone receptors (SHR) in the presence of ligand / RNA splicing / TBP-class protein binding / nuclear receptor binding / gluconeogenesis / respiratory electron transport chain / transcription coregulator activity / RNA polymerase II transcription regulatory region sequence-specific DNA binding / mitochondrion organization / synaptic transmission, glutamatergic / SUMOylation of intracellular receptors / transcription initiation at RNA polymerase II promoter / circadian regulation of gene expression / chromosome segregation / promoter-specific chromatin binding / negative regulation of smooth muscle cell proliferation / Heme signaling / Hsp90 protein binding / PPARA activates gene expression / Transcriptional activation of mitochondrial biogenesis / regulation of circadian rhythm / PML body / chromatin DNA binding / Nuclear Receptor transcription pathway / Transcriptional regulation of white adipocyte differentiation / response to wounding / positive regulation of miRNA transcription / DNA-binding transcription repressor activity, RNA polymerase II-specific / nuclear receptor activity / spindle / mRNA processing / sequence-specific double-stranded DNA binding / Regulation of RUNX2 expression and activity / : / positive regulation of neuron apoptotic process / positive regulation of cold-induced thermogenesis / gene expression / MLL4 and MLL3 complexes regulate expression of PPARG target genes in adipogenesis and hepatic steatosis / chromatin organization / protein-containing complex assembly / cellular response to oxidative stress / DNA-binding transcription activator activity, RNA polymerase II-specific / neuron apoptotic process / sequence-specific DNA binding / DNA-binding transcription factor binding / Potential therapeutics for SARS / negative regulation of neuron apoptotic process / RNA polymerase II-specific DNA-binding transcription factor binding / DNA-binding transcription factor activity, RNA polymerase II-specific / transcription coactivator activity / protein stabilization / nuclear speck / RNA polymerase II cis-regulatory region sequence-specific DNA binding / mitochondrial matrix / DNA-binding transcription factor activity / cell division / negative regulation of DNA-templated transcription / apoptotic process Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.7 Å MOLECULAR REPLACEMENT / Resolution: 2.7 Å | ||||||
 Authors Authors | Postel, S. / Edman, K. / Wissler, L. | ||||||
| Funding support | 1items
| ||||||
 Citation Citation |  Journal: Nat.Struct.Mol.Biol. / Year: 2023 Journal: Nat.Struct.Mol.Biol. / Year: 2023Title: Quaternary glucocorticoid receptor structure highlights allosteric interdomain communication. Authors: Postel, S. / Wissler, L. / Johansson, C.A. / Gunnarsson, A. / Gordon, E. / Collins, B. / Castaldo, M. / Kohler, C. / Oling, D. / Johansson, P. / Froderberg Roth, L. / Beinsteiner, B. / ...Authors: Postel, S. / Wissler, L. / Johansson, C.A. / Gunnarsson, A. / Gordon, E. / Collins, B. / Castaldo, M. / Kohler, C. / Oling, D. / Johansson, P. / Froderberg Roth, L. / Beinsteiner, B. / Dainty, I. / Delaney, S. / Klaholz, B.P. / Billas, I.M.L. / Edman, K. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7prv.cif.gz 7prv.cif.gz | 337.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7prv.ent.gz pdb7prv.ent.gz | 271.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7prv.json.gz 7prv.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/pr/7prv https://data.pdbj.org/pub/pdb/validation_reports/pr/7prv ftp://data.pdbj.org/pub/pdb/validation_reports/pr/7prv ftp://data.pdbj.org/pub/pdb/validation_reports/pr/7prv | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  7prwC  7prxC  3g9oS  4p6wS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-DNA chain , 2 types, 2 molecules CD
| #2: DNA chain | Mass: 7064.585 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.)  Homo sapiens (human) Homo sapiens (human) |
|---|---|
| #3: DNA chain | Mass: 7015.546 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.)  Homo sapiens (human) Homo sapiens (human) |
-Protein / Protein/peptide , 2 types, 3 molecules ABF
| #1: Protein | Mass: 44511.457 Da / Num. of mol.: 2 / Mutation: S404A, N517D, V571M, F602S, C638D Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: NR3C1, GRL / Cell line (production host): Sf21 / Production host: Homo sapiens (human) / Gene: NR3C1, GRL / Cell line (production host): Sf21 / Production host:  #4: Protein/peptide | | Mass: 2261.611 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.)  Homo sapiens (human) / References: UniProt: Q9UBK2 Homo sapiens (human) / References: UniProt: Q9UBK2 |
|---|
-Non-polymers , 4 types, 39 molecules 

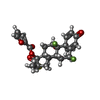




| #5: Chemical | ChemComp-ZN / #6: Chemical | #7: Chemical | #8: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.02 Å3/Da / Density % sol: 63.66 % |
|---|---|
| Crystal grow | Temperature: 277 K / Method: vapor diffusion, hanging drop / pH: 6.5 Details: 8 % PEG3350, 0.3 M 1,6-hexanediol, 0.1 M guanidine hydrochloride, 8 % 2,2,2-trifluoroethanol, 0.1 M Bis Tris Propane pH 6.5 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: MASSIF-1 / Wavelength: 0.96545 Å / Beamline: MASSIF-1 / Wavelength: 0.96545 Å |
| Detector | Type: DECTRIS PILATUS3 2M / Detector: PIXEL / Date: Mar 7, 2021 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.96545 Å / Relative weight: 1 |
| Reflection | Resolution: 2.7→68.684 Å / Num. obs: 24963 / % possible obs: 93.9 % / Redundancy: 6.5 % / CC1/2: 0.999 / Rmerge(I) obs: 0.067 / Rpim(I) all: 0.029 / Net I/σ(I): 16.3 |
| Reflection shell | Resolution: 2.7→3.005 Å / Redundancy: 6.9 % / Rmerge(I) obs: 1.035 / Mean I/σ(I) obs: 1.6 / Num. unique obs: 1248 / Rpim(I) all: 0.425 / % possible all: 63.5 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 3g9O, 4P6W Resolution: 2.7→68.68 Å / Cor.coef. Fo:Fc: 0.933 / Cor.coef. Fo:Fc free: 0.931 / Cross valid method: THROUGHOUT / SU Rfree Blow DPI: 0.382
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 103.55 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.4 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.7→68.68 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.7→2.88 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller


 PDBj
PDBj
















































