[English] 日本語
 Yorodumi
Yorodumi- PDB-7np8: Crystal structure of the Coenzyme F420-dependent sulfite reductas... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7np8 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Crystal structure of the Coenzyme F420-dependent sulfite reductase from Methanocaldococcus jannaschii at 2.3-A resolution | |||||||||
 Components Components | Coenzyme F420-dependent sulfite reductase | |||||||||
 Keywords Keywords | OXIDOREDUCTASE / Sulfite detoxification / Sulfur metabolism / sulfide / hydrogenotrophic methanogens / iron-sulfur cluster / sulfite-reductase evolution / sulfite/nitrite reductase / siroheme / flavin / ferredoxin / F420 / hyperthermophile | |||||||||
| Function / homology |  Function and homology information Function and homology informationsulfite reductase (coenzyme F420) / oxidoreductase activity, acting on CH or CH2 groups, with an iron-sulfur protein as acceptor / 4 iron, 4 sulfur cluster binding / heme binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |   Methanocaldococcus jannaschii DSM 2661 (archaea) Methanocaldococcus jannaschii DSM 2661 (archaea) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  SAD / Resolution: 2.3 Å SAD / Resolution: 2.3 Å | |||||||||
 Authors Authors | Jespersen, M. / Wagner, T. | |||||||||
| Funding support |  Germany, 2items Germany, 2items
| |||||||||
 Citation Citation |  Journal: Nat.Chem.Biol. / Year: 2023 Journal: Nat.Chem.Biol. / Year: 2023Title: Structures of the sulfite detoxifying F 420 -dependent enzyme from Methanococcales. Authors: Jespersen, M. / Pierik, A.J. / Wagner, T. #1:  Journal: Biorxiv / Year: 2022 Journal: Biorxiv / Year: 2022Title: The structure of the F420-dependent sulfite-detoxifying enzyme from Methanococcales reveals a prototypical sulfite-reductase with assimilatory traits Authors: Jespersen, M. / Pierik, A.J. / Wagner, T. | |||||||||
| History |
|
- Structure visualization
Structure visualization
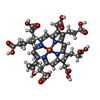
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7np8.cif.gz 7np8.cif.gz | 1011.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7np8.ent.gz pdb7np8.ent.gz | 844.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7np8.json.gz 7np8.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  7np8_validation.pdf.gz 7np8_validation.pdf.gz | 14.7 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  7np8_full_validation.pdf.gz 7np8_full_validation.pdf.gz | 14.8 MB | Display | |
| Data in XML |  7np8_validation.xml.gz 7np8_validation.xml.gz | 95.5 KB | Display | |
| Data in CIF |  7np8_validation.cif.gz 7np8_validation.cif.gz | 131.7 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/np/7np8 https://data.pdbj.org/pub/pdb/validation_reports/np/7np8 ftp://data.pdbj.org/pub/pdb/validation_reports/np/7np8 ftp://data.pdbj.org/pub/pdb/validation_reports/np/7np8 | HTTPS FTP |
-Related structure data
| Related structure data |  7npaC C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||||||||
| 2 | 
| ||||||||||||||||||
| Unit cell |
| ||||||||||||||||||
| Components on special symmetry positions |
|
- Components
Components
-Protein , 1 types, 4 molecules ABCD
| #1: Protein | Mass: 69902.047 Da / Num. of mol.: 4 / Source method: isolated from a natural source Source: (natural)   Methanocaldococcus jannaschii DSM 2661 (archaea) Methanocaldococcus jannaschii DSM 2661 (archaea)Cell line: / / Organ: / / Plasmid details: / / Variant: / / Tissue: / References: UniProt: Q58280, sulfite reductase (coenzyme F420) |
|---|
-Non-polymers , 10 types, 861 molecules 

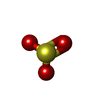







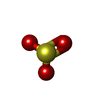







| #2: Chemical | ChemComp-SF4 / #3: Chemical | ChemComp-FAD / #4: Chemical | ChemComp-SO3 / #5: Chemical | ChemComp-SRM / #6: Chemical | ChemComp-GOL / #7: Chemical | ChemComp-MPD / ( #8: Chemical | ChemComp-CA / #9: Chemical | ChemComp-CL / #10: Chemical | ChemComp-FE / | #11: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.53 Å3/Da / Density % sol: 51.34 % / Description: Black, long plate-shaped crystals |
|---|---|
| Crystal grow | Temperature: 293.15 K / Method: vapor diffusion, sitting drop / pH: 5.5 Details: Purified Fsr from Methanocaldococcus jannaschii was crystallized anaerobically under yellow light at a concentration of 6.1 mg ml-1 in 25 mM Tris/HCl pH 7.6, 10% v/v glycerol and 2 mM ...Details: Purified Fsr from Methanocaldococcus jannaschii was crystallized anaerobically under yellow light at a concentration of 6.1 mg ml-1 in 25 mM Tris/HCl pH 7.6, 10% v/v glycerol and 2 mM dithiothreitol. 0.5 ul protein sample was mixed with 0.5 ul reservoir of crystallization solution containing 45% v/v 2-Methyl-2,4-pentandiol, 100 mM Bis-Tris pH 5.5 and 200 mM Calcium chloride. Crystals appeared after a few days. PH range: / / Temp details: +/- 1 degree of oscillation |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SOLEIL SOLEIL  / Beamline: PROXIMA 1 / Wavelength: 0.97857 Å / Beamline: PROXIMA 1 / Wavelength: 0.97857 Å |
| Detector | Type: DECTRIS EIGER X 16M / Detector: PIXEL / Date: Nov 22, 2019 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.97857 Å / Relative weight: 1 |
| Reflection twin | Operator: -k,-h,-l / Fraction: 0.05 |
| Reflection | Resolution: 2.299→78.823 Å / Num. obs: 104064 / % possible obs: 96 % / Redundancy: 13.9 % / CC1/2: 0.995 / Rmerge(I) obs: 0.251 / Rpim(I) all: 0.07 / Rrim(I) all: 0.261 / Net I/σ(I): 8.3 |
| Reflection shell | Resolution: 2.3→2.41 Å / Redundancy: 13.2 % / Rmerge(I) obs: 1.623 / Mean I/σ(I) obs: 1.6 / Num. unique obs: 5203 / CC1/2: 0.629 / Rpim(I) all: 0.459 / Rrim(I) all: 1.688 / % possible all: 94.8 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  SAD / Resolution: 2.3→78.82 Å / Cross valid method: THROUGHOUT / σ(F): 1.35 / Phase error: 22.55 / Stereochemistry target values: TWIN_LSQ_F SAD / Resolution: 2.3→78.82 Å / Cross valid method: THROUGHOUT / σ(F): 1.35 / Phase error: 22.55 / Stereochemistry target values: TWIN_LSQ_FDetails: Refinement was performed with the twin law:-k,-h,-l
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 139.85 Å2 / Biso mean: 42.4377 Å2 / Biso min: 16.19 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 2.3→78.82 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Rfactor Rfree error: 0 / Total num. of bins used: 20
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller


 PDBj
PDBj
















