+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7lna | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Infectious mammalian prion fibril (263K scrapie) | |||||||||
 Components Components | Major prion protein | |||||||||
 Keywords Keywords | PROTEIN FIBRIL / infectious mammalian prion / templating / glycosylated glycophophatidlyinositol-anchored amyloid / PIRIBS | |||||||||
| Function / homology |  Function and homology information Function and homology informationaspartic-type endopeptidase inhibitor activity / positive regulation of glutamate receptor signaling pathway / regulation of calcium ion import across plasma membrane / glycosaminoglycan binding / type 5 metabotropic glutamate receptor binding / negative regulation of interleukin-17 production / cupric ion binding / regulation of potassium ion transmembrane transport / negative regulation of dendritic spine maintenance / negative regulation of calcineurin-NFAT signaling cascade ...aspartic-type endopeptidase inhibitor activity / positive regulation of glutamate receptor signaling pathway / regulation of calcium ion import across plasma membrane / glycosaminoglycan binding / type 5 metabotropic glutamate receptor binding / negative regulation of interleukin-17 production / cupric ion binding / regulation of potassium ion transmembrane transport / negative regulation of dendritic spine maintenance / negative regulation of calcineurin-NFAT signaling cascade / negative regulation of interleukin-2 production / negative regulation of T cell receptor signaling pathway / negative regulation of activated T cell proliferation / negative regulation of amyloid-beta formation / negative regulation of type II interferon production / cuprous ion binding / positive regulation of protein targeting to membrane / side of membrane / inclusion body / neuron projection maintenance / positive regulation of calcium-mediated signaling / molecular function activator activity / cellular response to copper ion / positive regulation of protein localization to plasma membrane / molecular condensate scaffold activity / protein homooligomerization / protein destabilization / cellular response to xenobiotic stimulus / cellular response to amyloid-beta / terminal bouton / positive regulation of neuron apoptotic process / signaling receptor activity / presynapse / amyloid-beta binding / response to oxidative stress / protease binding / nuclear membrane / microtubule binding / amyloid fibril formation / learning or memory / regulation of cell cycle / intracellular signal transduction / membrane raft / copper ion binding / dendrite / negative regulation of apoptotic process / protein-containing complex binding / cell surface / negative regulation of transcription by RNA polymerase II / endoplasmic reticulum / Golgi apparatus / identical protein binding / plasma membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  Mesocricetus auratus (golden hamster) Mesocricetus auratus (golden hamster) | |||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.14 Å | |||||||||
 Authors Authors | Kraus, A. / Hoyt, F. / Schwartz, C.L. / Hansen, B. / Hughson, A.G. / Artikis, E. / Race, B. / Caughey, B. | |||||||||
| Funding support |  United States, 2items United States, 2items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2021 Journal: Mol Cell / Year: 2021Title: High-resolution structure and strain comparison of infectious mammalian prions. Authors: Allison Kraus / Forrest Hoyt / Cindi L Schwartz / Bryan Hansen / Efrosini Artikis / Andrew G Hughson / Gregory J Raymond / Brent Race / Gerald S Baron / Byron Caughey /  Abstract: Within the extensive range of self-propagating pathologic protein aggregates of mammals, prions are the most clearly infectious (e.g., ∼10 lethal doses per milligram). The structures of such lethal ...Within the extensive range of self-propagating pathologic protein aggregates of mammals, prions are the most clearly infectious (e.g., ∼10 lethal doses per milligram). The structures of such lethal assemblies of PrP molecules have been poorly understood. Here we report a near-atomic core structure of a brain-derived, fully infectious prion (263K strain). Cryo-electron microscopy showed amyloid fibrils assembled with parallel in-register intermolecular β sheets. Each monomer provides one rung of the ordered fibril core, with N-linked glycans and glycolipid anchors projecting outward. Thus, single monomers form the templating surface for incoming monomers at fibril ends, where prion growth occurs. Comparison to another prion strain (aRML) revealed major differences in fibril morphology but, like 263K, an asymmetric fibril cross-section without paired protofilaments. These findings provide structural insights into prion propagation, strains, species barriers, and membrane pathogenesis. This structure also helps frame considerations of factors influencing the relative transmissibility of other pathologic amyloids. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7lna.cif.gz 7lna.cif.gz | 80.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7lna.ent.gz pdb7lna.ent.gz | 56.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7lna.json.gz 7lna.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ln/7lna https://data.pdbj.org/pub/pdb/validation_reports/ln/7lna ftp://data.pdbj.org/pub/pdb/validation_reports/ln/7lna ftp://data.pdbj.org/pub/pdb/validation_reports/ln/7lna | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  23459MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 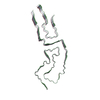
| |||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
NCS domain segments: Component-ID: _ / Beg auth comp-ID: THR / Beg label comp-ID: THR / End auth comp-ID: ASP / End label comp-ID: ASP / Refine code: _ / Auth seq-ID: 95 - 227 / Label seq-ID: 6 - 138
NCS ensembles :
|
- Components
Components
| #1: Protein | Mass: 16264.101 Da / Num. of mol.: 3 / Source method: isolated from a natural source Details: Protease-resistant glycosylated, glycophophatidlyinositol-anchored infectious prion amyloid core (inclusive of residues 90-231). Source: (natural)  Mesocricetus auratus (golden hamster) / Organ: brain / References: UniProt: P04273 Mesocricetus auratus (golden hamster) / Organ: brain / References: UniProt: P04273Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: FILAMENT / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: infectious mammalian prion fibril (263K scrapie) / Type: COMPLEX Details: Purified from brains of hamsters with clinical scrapie prion disease. Entity ID: all / Source: NATURAL |
|---|---|
| Molecular weight | Experimental value: NO |
| Source (natural) | Organism:  Mesocricetus auratus (golden hamster) / Organ: brain / Tissue: brain Mesocricetus auratus (golden hamster) / Organ: brain / Tissue: brain |
| Buffer solution | pH: 7.4 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Specimen support | Details: Solarus 950 (Gatan, Pleasanton CA) / Grid material: COPPER / Grid mesh size: 300 divisions/in. / Grid type: C-flat-1.2/1.3 |
| Vitrification | Instrument: LEICA EM GP / Cryogen name: ETHANE / Humidity: 90 % / Chamber temperature: 295 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: OTHER FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: OTHER |
| Electron lens | Mode: BRIGHT FIELD |
| Specimen holder | Cryogen: NITROGEN |
| Image recording | Electron dose: 60 e/Å2 / Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) |
- Processing
Processing
| Software | Name: REFMAC / Version: 5.8.0258 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Image processing | Details: Processing and reconstruction was conducted in Relion as per Scheres, Acta Cryst. (2020). D76, 94-101 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 337368 Details: Filament start and end positions were picked manually in RELION. Particles were extracted with an interbox distance of 14.7A along the filament axis. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Point symmetry: C1 (asymmetric) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3.14 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 15884 / Algorithm: FOURIER SPACE Details: Auto-refinement was then performed while optimizing the helical twist and rise. Auto-refinement with refinement of twist and rise yielded a final map with a twist of -0.847 degree and rise ...Details: Auto-refinement was then performed while optimizing the helical twist and rise. Auto-refinement with refinement of twist and rise yielded a final map with a twist of -0.847 degree and rise of 4.874 angstrom. Iterative cycles of CTF refinement, Bayesian polishing, and auto refinement were used until resolution estimates stabilized. Post processing in RELION was performed with a soft-edged mask representing 10% of the central Z length of the fibril. Sharpening was applied with a B-factor of -31 square angstrom. Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Atomic model building | Protocol: AB INITIO MODEL / Space: RECIPROCAL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | Resolution: 3.14→3.14 Å / Cor.coef. Fo:Fc: 0.808 / ESU R: 0.544 Stereochemistry target values: MAXIMUM LIKELIHOOD WITH PHASES
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: PARAMETERS FOR MASK CACLULATION | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 39.924 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: 1 / Total: 3171 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller






 PDBj
PDBj
