[English] 日本語
 Yorodumi
Yorodumi- PDB-7kjx: Structure of HIV-1 reverse transcriptase initiation complex core ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7kjx | ||||||
|---|---|---|---|---|---|---|---|
| Title | Structure of HIV-1 reverse transcriptase initiation complex core with nevirapine | ||||||
 Components Components |
| ||||||
 Keywords Keywords | viral protein/RNA / reverse transcriptase / RNA / protein-RNA complex / tRNA / polymerase / viral protein / viral protein-RNA complex | ||||||
| Function / homology |  Function and homology information Function and homology informationHIV-1 retropepsin / symbiont-mediated activation of host apoptosis / retroviral ribonuclease H / exoribonuclease H / exoribonuclease H activity / DNA integration / viral genome integration into host DNA / establishment of integrated proviral latency / RNA-directed DNA polymerase / RNA stem-loop binding ...HIV-1 retropepsin / symbiont-mediated activation of host apoptosis / retroviral ribonuclease H / exoribonuclease H / exoribonuclease H activity / DNA integration / viral genome integration into host DNA / establishment of integrated proviral latency / RNA-directed DNA polymerase / RNA stem-loop binding / viral penetration into host nucleus / host multivesicular body / RNA-directed DNA polymerase activity / RNA-DNA hybrid ribonuclease activity / Transferases; Transferring phosphorus-containing groups; Nucleotidyltransferases / host cell / viral nucleocapsid / DNA recombination / DNA-directed DNA polymerase / aspartic-type endopeptidase activity / Hydrolases; Acting on ester bonds / DNA-directed DNA polymerase activity / symbiont-mediated suppression of host gene expression / viral translational frameshifting / symbiont entry into host cell / lipid binding / host cell nucleus / host cell plasma membrane / virion membrane / structural molecule activity / proteolysis / DNA binding / zinc ion binding Similarity search - Function | ||||||
| Biological species |  Human immunodeficiency virus type 1 group M subtype B Human immunodeficiency virus type 1 group M subtype B  Human immunodeficiency virus 1 Human immunodeficiency virus 1 Homo sapiens (human) Homo sapiens (human) | ||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.1 Å | ||||||
 Authors Authors | Ha, B. / Larsen, K.P. / Zhang, J. / Fu, Z. / Montabana, E. / Jackson, L.N. / Chen, D.H. / Puglisi, E.V. | ||||||
| Funding support |  United States, 1items United States, 1items
| ||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: High-resolution view of HIV-1 reverse transcriptase initiation complexes and inhibition by NNRTI drugs. Authors: Betty Ha / Kevin P Larsen / Jingji Zhang / Ziao Fu / Elizabeth Montabana / Lynnette N Jackson / Dong-Hua Chen / Elisabetta Viani Puglisi /  Abstract: Reverse transcription of the HIV-1 viral RNA genome (vRNA) is an integral step in virus replication. Upon viral entry, HIV-1 reverse transcriptase (RT) initiates from a host tRNA primer bound to the ...Reverse transcription of the HIV-1 viral RNA genome (vRNA) is an integral step in virus replication. Upon viral entry, HIV-1 reverse transcriptase (RT) initiates from a host tRNA primer bound to the vRNA genome and is the target of key antivirals, such as non-nucleoside reverse transcriptase inhibitors (NNRTIs). Initiation proceeds slowly with discrete pausing events along the vRNA template. Despite prior medium-resolution structural characterization of reverse transcriptase initiation complexes (RTICs), higher-resolution structures of the RTIC are needed to understand the molecular mechanisms that underlie initiation. Here we report cryo-EM structures of the core RTIC, RTIC-nevirapine, and RTIC-efavirenz complexes at 2.8, 3.1, and 2.9 Å, respectively. In combination with biochemical studies, these data suggest a basis for rapid dissociation kinetics of RT from the vRNA-tRNA initiation complex and reveal a specific structural mechanism of nucleic acid conformational stabilization during initiation. Finally, our results show that NNRTIs inhibit the RTIC and exacerbate discrete pausing during early reverse transcription. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7kjx.cif.gz 7kjx.cif.gz | 205.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7kjx.ent.gz pdb7kjx.ent.gz | 156.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7kjx.json.gz 7kjx.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/kj/7kjx https://data.pdbj.org/pub/pdb/validation_reports/kj/7kjx ftp://data.pdbj.org/pub/pdb/validation_reports/kj/7kjx ftp://data.pdbj.org/pub/pdb/validation_reports/kj/7kjx | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  22901MC  7kjvC  7kjwC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Protein , 2 types, 2 molecules AB
| #1: Protein | Mass: 64705.316 Da / Num. of mol.: 1 / Mutation: Q258C, E478Q Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Human immunodeficiency virus type 1 group M subtype B (isolate BH10) Human immunodeficiency virus type 1 group M subtype B (isolate BH10)Strain: isolate BH10 / Gene: gag-pol / Production host:  References: UniProt: P03366, RNA-directed DNA polymerase, DNA-directed DNA polymerase, retroviral ribonuclease H |
|---|---|
| #2: Protein | Mass: 51585.293 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Human immunodeficiency virus 1 / Strain: isolate BH10 / Gene: gag-pol / Production host: Human immunodeficiency virus 1 / Strain: isolate BH10 / Gene: gag-pol / Production host:  |
-RNA chain / DNA/RNA hybrid , 2 types, 2 molecules CD
| #3: RNA chain | Mass: 8437.105 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.)   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
|---|---|
| #4: DNA/RNA hybrid | Mass: 7030.295 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.)  Homo sapiens (human) Homo sapiens (human) |
-Non-polymers , 2 types, 2 molecules 
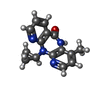

| #5: Chemical | ChemComp-MG / |
|---|---|
| #6: Chemical | ChemComp-NVP / |
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: HIV-1 reverse transcriptase initiation complex core with nevirapine Type: COMPLEX Details: Peripheral tRNA and engineered stem loop were masked out during cryo-EM data processing. Entity ID: #1-#4 / Source: MULTIPLE SOURCES |
|---|---|
| Molecular weight | Value: 0.138 MDa / Experimental value: NO |
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Buffer solution | pH: 8 |
| Specimen | Conc.: 10 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES / Details: Sample was monodisperse |
| Specimen support | Grid material: GOLD / Grid mesh size: 200 divisions/in. / Grid type: Quantifoil R0.6/1 |
| Vitrification | Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 295.15 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD |
| Image recording | Electron dose: 100 e/Å2 / Film or detector model: GATAN K2 SUMMIT (4k x 4k) |
- Processing
Processing
| EM software |
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | |||||||||||||||
| 3D reconstruction | Resolution: 3.1 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 1155315 / Symmetry type: POINT | |||||||||||||||
| Atomic model building | Space: REAL |
 Movie
Movie Controller
Controller














 PDBj
PDBj

































