+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: PDB / ID: 7ke6 | ||||||
|---|---|---|---|---|---|---|---|
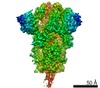
| タイトル | SARS-CoV-2 D614G 3 RBD down Spike Protein Trimer without the P986-P987 stabilizing mutations (S-GSAS-D614G sub-classification) | ||||||
 要素 要素 | Spike glycoprotein | ||||||
 キーワード キーワード | VIRAL PROTEIN / SARS-CoV-2 Spike Protein Trimer | ||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / viral translation / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / viral translation / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / entry receptor-mediated virion attachment to host cell / membrane fusion / Attachment and Entry / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / receptor ligand activity / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / identical protein binding / membrane / plasma membrane 類似検索 - 分子機能 | ||||||
| 生物種 |  | ||||||
| 手法 | 電子顕微鏡法 / 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 3.1 Å | ||||||
 データ登録者 データ登録者 | Gobeil, S. / Acharya, P. | ||||||
| 資金援助 |  米国, 1件 米国, 1件
| ||||||
 引用 引用 |  ジャーナル: Cell Rep / 年: 2021 ジャーナル: Cell Rep / 年: 2021タイトル: D614G Mutation Alters SARS-CoV-2 Spike Conformation and Enhances Protease Cleavage at the S1/S2 Junction. 著者: Sophie M-C Gobeil / Katarzyna Janowska / Shana McDowell / Katayoun Mansouri / Robert Parks / Kartik Manne / Victoria Stalls / Megan F Kopp / Rory Henderson / Robert J Edwards / Barton F ...著者: Sophie M-C Gobeil / Katarzyna Janowska / Shana McDowell / Katayoun Mansouri / Robert Parks / Kartik Manne / Victoria Stalls / Megan F Kopp / Rory Henderson / Robert J Edwards / Barton F Haynes / Priyamvada Acharya /  要旨: The severe acute respiratory coronavirus 2 (SARS-CoV-2) spike (S) protein is the target of vaccine design efforts to end the coronavirus disease 2019 (COVID-19) pandemic. Despite a low mutation rate, ...The severe acute respiratory coronavirus 2 (SARS-CoV-2) spike (S) protein is the target of vaccine design efforts to end the coronavirus disease 2019 (COVID-19) pandemic. Despite a low mutation rate, isolates with the D614G substitution in the S protein appeared early during the pandemic and are now the dominant form worldwide. Here, we explore S conformational changes and the effects of the D614G mutation on a soluble S ectodomain construct. Cryoelectron microscopy (cryo-EM) structures reveal altered receptor binding domain (RBD) disposition; antigenicity and proteolysis experiments reveal structural changes and enhanced furin cleavage efficiency of the G614 variant. Furthermore, furin cleavage alters the up/down ratio of the RBDs in the G614 S ectodomain, demonstrating an allosteric effect on RBD positioning triggered by changes in the SD2 region, which harbors residue 614 and the furin cleavage site. Our results elucidate SARS-CoV-2 S conformational landscape and allostery and have implications for vaccine design. #1: ジャーナル: bioRxiv / 年: 2020 タイトル: D614G mutation alters SARS-CoV-2 spike conformational dynamics and protease cleavage susceptibility at the S1/S2 junction. 著者: Sophie Gobeil / Katarzyna Janowska / Shana McDowell / Katayoun Mansouri / Robert Parks / Kartik Manne / Victoria Stalls / Megan Kopp / Rory Henderson / Robert J Edwards / Barton F Haynes / Priyamvada Acharya 要旨: The SARS-CoV-2 spike (S) protein is the target of vaccine design efforts to end the COVID-19 pandemic. Despite a low mutation rate, isolates with the D614G substitution in the S protein appeared ...The SARS-CoV-2 spike (S) protein is the target of vaccine design efforts to end the COVID-19 pandemic. Despite a low mutation rate, isolates with the D614G substitution in the S protein appeared early during the pandemic, and are now the dominant form worldwide. Here, we analyze the D614G mutation in the context of a soluble S ectodomain construct. Cryo-EM structures, antigenicity and proteolysis experiments suggest altered conformational dynamics resulting in enhanced furin cleavage efficiency of the G614 variant. Furthermore, furin cleavage alters the conformational dynamics of the Receptor Binding Domains (RBD) in the G614 S ectodomain, demonstrating an allosteric effect on the RBD dynamics triggered by changes in the SD2 region, that harbors residue 614 and the furin cleavage site. Our results elucidate SARS-CoV-2 spike conformational dynamics and allostery, and have implications for vaccine design. | ||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | 分子:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- ダウンロードとリンク
ダウンロードとリンク
- ダウンロード
ダウンロード
| PDBx/mmCIF形式 |  7ke6.cif.gz 7ke6.cif.gz | 599.3 KB | 表示 |  PDBx/mmCIF形式 PDBx/mmCIF形式 |
|---|---|---|---|---|
| PDB形式 |  pdb7ke6.ent.gz pdb7ke6.ent.gz | 476.3 KB | 表示 |  PDB形式 PDB形式 |
| PDBx/mmJSON形式 |  7ke6.json.gz 7ke6.json.gz | ツリー表示 |  PDBx/mmJSON形式 PDBx/mmJSON形式 | |
| その他 |  その他のダウンロード その他のダウンロード |
-検証レポート
| 文書・要旨 |  7ke6_validation.pdf.gz 7ke6_validation.pdf.gz | 1.8 MB | 表示 |  wwPDB検証レポート wwPDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  7ke6_full_validation.pdf.gz 7ke6_full_validation.pdf.gz | 1.8 MB | 表示 | |
| XML形式データ |  7ke6_validation.xml.gz 7ke6_validation.xml.gz | 72.4 KB | 表示 | |
| CIF形式データ |  7ke6_validation.cif.gz 7ke6_validation.cif.gz | 114.9 KB | 表示 | |
| アーカイブディレクトリ |  https://data.pdbj.org/pub/pdb/validation_reports/ke/7ke6 https://data.pdbj.org/pub/pdb/validation_reports/ke/7ke6 ftp://data.pdbj.org/pub/pdb/validation_reports/ke/7ke6 ftp://data.pdbj.org/pub/pdb/validation_reports/ke/7ke6 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  22832MC  7kdgC  7kdhC  7kdiC  7kdjC  7kdkC  7kdlC  7ke4C  7ke7C  7ke8C  7ke9C  7keaC  7kebC  7kecC M: このデータのモデリングに利用したマップデータ C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ |
- リンク
リンク
- 集合体
集合体
| 登録構造単位 | 
|
|---|---|
| 1 |
|
- 要素
要素
| #1: タンパク質 | 分子量: 142375.422 Da / 分子数: 3 / 変異: D614G R682G R683S R685S / 由来タイプ: 組換発現 由来: (組換発現)  遺伝子: S, 2 / 発現宿主:  Homo sapiens (ヒト) / 参照: UniProt: P0DTC2 Homo sapiens (ヒト) / 参照: UniProt: P0DTC2#2: 多糖 | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose #3: 糖 | ChemComp-NAG / 研究の焦点であるリガンドがあるか | N | Has protein modification | Y | |
|---|
-実験情報
-実験
| 実験 | 手法: 電子顕微鏡法 |
|---|---|
| EM実験 | 試料の集合状態: PARTICLE / 3次元再構成法: 単粒子再構成法 |
- 試料調製
試料調製
| 構成要素 | 名称: Spike protein Trimer / タイプ: COMPLEX / Entity ID: #1 / 由来: RECOMBINANT |
|---|---|
| 由来(天然) | 生物種:  |
| 由来(組換発現) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 緩衝液 | pH: 8 |
| 試料 | 包埋: NO / シャドウイング: NO / 染色: NO / 凍結: YES |
| 急速凍結 | 凍結剤: ETHANE |
- 電子顕微鏡撮影
電子顕微鏡撮影
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
|---|---|
| 顕微鏡 | モデル: FEI TITAN KRIOS |
| 電子銃 | 電子線源:  FIELD EMISSION GUN / 加速電圧: 300 kV / 照射モード: OTHER FIELD EMISSION GUN / 加速電圧: 300 kV / 照射モード: OTHER |
| 電子レンズ | モード: BRIGHT FIELD |
| 撮影 | 電子線照射量: 51.8 e/Å2 / フィルム・検出器のモデル: GATAN K3 (6k x 4k) |
- 解析
解析
| CTF補正 | タイプ: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3次元再構成 | 解像度: 3.1 Å / 解像度の算出法: FSC 0.143 CUT-OFF / 粒子像の数: 196173 / 対称性のタイプ: POINT | ||||||||||||||||||||||||
| 精密化 | 立体化学のターゲット値: GeoStd + Monomer Library + CDL v1.2 | ||||||||||||||||||||||||
| 原子変位パラメータ | Biso mean: 28.72 Å2 | ||||||||||||||||||||||||
| 拘束条件 |
|
 ムービー
ムービー コントローラー
コントローラー

























 PDBj
PDBj





