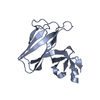
Entry Database : PDB / ID : 7ehaTitle Crystal structure of the flagellar hook cap from Salmonella enterica serovar Typhimurium Basal-body rod modification protein FlgD Keywords / / / / Function / homology / / / / / / / Biological species Salmonella typhimurium (bacteria)Method / / / Resolution : 3.3 Å Authors Matsunami, H. / Yoon, Y.-H. / Imada, K. / Namba, K. / Samatey, F.A. Funding support Organization Grant number Country Japan Society for the Promotion of Science (JSPS)
Journal : Commun Biol / Year : 2021Title : Structure of the bacterial flagellar hook cap provides insights into a hook assembly mechanismAuthors : Matsunami, H. / Yoon, Y.H. / Imada, K. / Namba, K. / Samatey, F.A. History Deposition Mar 29, 2021 Deposition site / Processing site Supersession Nov 24, 2021 ID 6IEE Revision 1.0 Nov 24, 2021 Provider / Type Revision 1.1 May 29, 2024 Group / Refinement descriptionCategory / chem_comp_bond / struct_ncs_dom_limItem _struct_ncs_dom_lim.beg_auth_comp_id / _struct_ncs_dom_lim.beg_label_asym_id ... _struct_ncs_dom_lim.beg_auth_comp_id / _struct_ncs_dom_lim.beg_label_asym_id / _struct_ncs_dom_lim.beg_label_comp_id / _struct_ncs_dom_lim.beg_label_seq_id / _struct_ncs_dom_lim.end_auth_comp_id / _struct_ncs_dom_lim.end_label_asym_id / _struct_ncs_dom_lim.end_label_comp_id / _struct_ncs_dom_lim.end_label_seq_id / _struct_ncs_dom_lim.selection_details
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Salmonella typhimurium (bacteria)
Salmonella typhimurium (bacteria) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MAD / Resolution: 3.3 Å
MAD / Resolution: 3.3 Å  Authors
Authors Japan, 1items
Japan, 1items  Citation
Citation Journal: Commun Biol / Year: 2021
Journal: Commun Biol / Year: 2021 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 7eha.cif.gz
7eha.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb7eha.ent.gz
pdb7eha.ent.gz PDB format
PDB format 7eha.json.gz
7eha.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/eh/7eha
https://data.pdbj.org/pub/pdb/validation_reports/eh/7eha ftp://data.pdbj.org/pub/pdb/validation_reports/eh/7eha
ftp://data.pdbj.org/pub/pdb/validation_reports/eh/7eha Links
Links Assembly
Assembly
 Movie
Movie Controller
Controller










 PDBj
PDBj