[English] 日本語
 Yorodumi
Yorodumi- PDB-7efd: 1.77 A cryo-EM structure of Streptavidin using first 40 frames (c... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7efd | ||||||
|---|---|---|---|---|---|---|---|
| Title | 1.77 A cryo-EM structure of Streptavidin using first 40 frames (corresponding to about 40 e/A^2 total dose) | ||||||
 Components Components | Streptavidin | ||||||
 Keywords Keywords | CYTOSOLIC PROTEIN / STREPTAVIDIN | ||||||
| Function / homology |  Function and homology information Function and homology information | ||||||
| Biological species |  Streptomyces avidinii (bacteria) Streptomyces avidinii (bacteria) | ||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 1.77 Å | ||||||
 Authors Authors | Hiraizumi, M. / Yamashita, K. / Nishizawa, T. / Kotecha, A. / Nureki, O. | ||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: 1.77 A cryo-EM structure of Streptavidin using first 40 frames (corresponding to about 40e/A^2 total dose) Authors: Hiraizumi, M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7efd.cif.gz 7efd.cif.gz | 42.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7efd.ent.gz pdb7efd.ent.gz | 24.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7efd.json.gz 7efd.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ef/7efd https://data.pdbj.org/pub/pdb/validation_reports/ef/7efd ftp://data.pdbj.org/pub/pdb/validation_reports/ef/7efd ftp://data.pdbj.org/pub/pdb/validation_reports/ef/7efd | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  31084MC  7efcC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||||||||||
| 2 |
| ||||||||||||||||||||
| 3 | 
| ||||||||||||||||||||
| Symmetry | Point symmetry: (Schoenflies symbol: D2 (2x2 fold dihedral)) | ||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS oper:
|
- Components
Components
| #1: Protein | Mass: 18849.672 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Streptomyces avidinii (bacteria) / References: UniProt: P22629 Streptomyces avidinii (bacteria) / References: UniProt: P22629 |
|---|---|
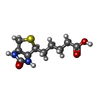
| #2: Chemical | ChemComp-BTN / |
| #3: Water | ChemComp-HOH / |
| Has ligand of interest | N |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Streptavidin / Type: COMPLEX / Entity ID: #1 / Source: NATURAL |
|---|---|
| Source (natural) | Organism:  Streptomyces avidinii (bacteria) Streptomyces avidinii (bacteria) |
| Buffer solution | pH: 7.2 |
| Specimen | Conc.: 1 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Specimen support | Grid material: GOLD / Grid mesh size: 300 divisions/in. / Grid type: UltrAuFoil R1.2/1.3 |
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: TFS KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD |
| Image recording | Electron dose: 70 e/Å2 / Film or detector model: FEI FALCON IV (4k x 4k) / Num. of grids imaged: 1 / Num. of real images: 4053 |
- Processing
Processing
| Software | Name: REFMAC / Version: 5.8.0272 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Point symmetry: D2 (2x2 fold dihedral) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 1.77 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 153976 / Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Atomic model building | Space: RECIPROCAL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Atomic model building | 3D fitting-ID: 1 / Source name: PDB / Type: experimental model
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | Resolution: 1.77→76.255 Å / Cor.coef. Fo:Fc: 0.895 / WRfactor Rwork: 0.349 / SU B: 3.828 / SU ML: 0.099 / Average fsc overall: 0.6978 / Average fsc work: 0.6978 / ESU R: 0.053 Details: Hydrogens have been added in their riding positions
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: BABINET MODEL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 45.492 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller













 PDBj
PDBj