+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7dij | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Falcilysin in complex with MK-4815 | |||||||||
 Components Components | Falcilysin | |||||||||
 Keywords Keywords | PROTEIN BINDING/INHIBITOR / PROTEIN BINDING-inhibitor complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationhemoglobin catabolic process / apicoplast / food vacuole / Hydrolases; Acting on peptide bonds (peptidases); Metalloendopeptidases / vacuolar membrane / metalloendopeptidase activity / protein processing / metal ion binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.9 Å MOLECULAR REPLACEMENT / Resolution: 1.9 Å | |||||||||
 Authors Authors | Lin, J.Q. / El Sahili, A. / Lescar, J. | |||||||||
| Funding support | 1items
| |||||||||
 Citation Citation |  Journal: Cell Chem Biol / Year: 2024 Journal: Cell Chem Biol / Year: 2024Title: Identification of an inhibitory pocket in falcilysin provides a new avenue for malaria drug development. Authors: Wirjanata, G. / Lin, J. / Dziekan, J.M. / El Sahili, A. / Chung, Z. / Tjia, S. / Binte Zulkifli, N.E. / Boentoro, J. / Tham, R. / Jia, L.S. / Go, K.D. / Yu, H. / Partridge, A. / Olsen, D. / ...Authors: Wirjanata, G. / Lin, J. / Dziekan, J.M. / El Sahili, A. / Chung, Z. / Tjia, S. / Binte Zulkifli, N.E. / Boentoro, J. / Tham, R. / Jia, L.S. / Go, K.D. / Yu, H. / Partridge, A. / Olsen, D. / Prabhu, N. / Sobota, R.M. / Nordlund, P. / Lescar, J. / Bozdech, Z. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7dij.cif.gz 7dij.cif.gz | 289.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7dij.ent.gz pdb7dij.ent.gz | 202.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7dij.json.gz 7dij.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  7dij_validation.pdf.gz 7dij_validation.pdf.gz | 1.2 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  7dij_full_validation.pdf.gz 7dij_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  7dij_validation.xml.gz 7dij_validation.xml.gz | 48.4 KB | Display | |
| Data in CIF |  7dij_validation.cif.gz 7dij_validation.cif.gz | 75.3 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/di/7dij https://data.pdbj.org/pub/pdb/validation_reports/di/7dij ftp://data.pdbj.org/pub/pdb/validation_reports/di/7dij ftp://data.pdbj.org/pub/pdb/validation_reports/di/7dij | HTTPS FTP |
-Related structure data
| Related structure data |  7di7C  7diaC  8ho4C  8ho5C  3s5mS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 135039.359 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   References: UniProt: Q76NL8, Hydrolases; Acting on peptide bonds (peptidases); Metalloendopeptidases |
|---|
-Non-polymers , 7 types, 1027 molecules 



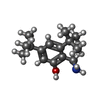








| #2: Chemical | ChemComp-GOL / | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| #3: Chemical | | #4: Chemical | #5: Chemical | ChemComp-DMS / | #6: Chemical | ChemComp-H8F / | #7: Chemical | ChemComp-ZN / | #8: Water | ChemComp-HOH / | |
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.48 Å3/Da / Density % sol: 50.48 % Description: THE ENTRY CONTAINS FRIEDEL PAIRS IN F_PLUS/MINUS COLUMNS. |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop Details: 0.2 M ammonium acetate, 0.1 M Tris pH 8.5, 25% w/v PEG 3,350 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SLS SLS  / Beamline: X06DA / Wavelength: 1 Å / Beamline: X06DA / Wavelength: 1 Å |
| Detector | Type: DECTRIS PILATUS 2M-F / Detector: PIXEL / Date: Nov 9, 2019 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 1.9→48.78 Å / Num. obs: 98800 / % possible obs: 99.85 % / Redundancy: 13.2 % / Biso Wilson estimate: 26.65 Å2 / CC1/2: 0.999 / CC star: 1 / Rmerge(I) obs: 0.1433 / Rpim(I) all: 0.04084 / Rrim(I) all: 0.1491 / Net I/σ(I): 15.48 |
| Reflection shell | Resolution: 1.9→1.968 Å / Redundancy: 13.1 % / Rmerge(I) obs: 1.358 / Num. unique obs: 9660 / CC1/2: 0.738 / CC star: 0.922 / Rpim(I) all: 0.3854 / % possible all: 98.83 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 3S5M Resolution: 1.9→48.78 Å / SU ML: 0.235 / Cross valid method: FREE R-VALUE / σ(F): 1.34 / Phase error: 22.2796 Stereochemistry target values: GeoStd + Monomer Library + CDL v1.2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 30.68 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.9→48.78 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller













 PDBj
PDBj







