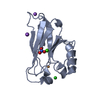
Entry Database : PDB / ID : 7b4uTitle Broadly neutralizing DARPin bnD.2 in complex with the HIV-1 envelope variable loop 3 crown mimetic peptide V3-IF (BG505) Broadly neutralizing DARPin bnD.2 HIV-1 envelope variable loop 3 crown mimetic peptide V3-IF (BG505) Keywords / / / / / / Biological species synthetic construct (others) Method / / / / Resolution : 1.45 Å Authors Friedrich, N. / Stiegeler, E. / Glogl, M. / Lemmin, T. / Hansen, S. / Kadelka, C. / Wu, Y. / Ernst, P. / Maliqi, L. / Foulkes, C. ...Friedrich, N. / Stiegeler, E. / Glogl, M. / Lemmin, T. / Hansen, S. / Kadelka, C. / Wu, Y. / Ernst, P. / Maliqi, L. / Foulkes, C. / Morin, M. / Eroglu, M. / Liechti, T. / Ivan, B. / Reinberg, T. / Schaefer, J. / Karakus, U. / Ursprung, S. / Mann, A. / Rusert, P. / Kouyos, R.D. / Robinson, J.A. / Gunthard, H.F. / Pluckthun, A. / Trkola, A. Funding support Organization Grant number Country Swiss National Science Foundation 310030_192689 Swiss National Science Foundation 310030B_166676 Swiss National Science Foundation 31003A_146278
Journal : Nat Commun / Year : 2021Title : Distinct conformations of the HIV-1 V3 loop crown are targetable for broad neutralization.Authors: Friedrich, N. / Stiegeler, E. / Glogl, M. / Lemmin, T. / Hansen, S. / Kadelka, C. / Wu, Y. / Ernst, P. / Maliqi, L. / Foulkes, C. / Morin, M. / Eroglu, M. / Liechti, T. / Ivan, B. / ... Authors : Friedrich, N. / Stiegeler, E. / Glogl, M. / Lemmin, T. / Hansen, S. / Kadelka, C. / Wu, Y. / Ernst, P. / Maliqi, L. / Foulkes, C. / Morin, M. / Eroglu, M. / Liechti, T. / Ivan, B. / Reinberg, T. / Schaefer, J.V. / Karakus, U. / Ursprung, S. / Mann, A. / Rusert, P. / Kouyos, R.D. / Robinson, J.A. / Gunthard, H.F. / Pluckthun, A. / Trkola, A. History Deposition Dec 2, 2020 Deposition site / Processing site Revision 1.0 Nov 24, 2021 Provider / Type Revision 1.1 Jun 15, 2022 Group / Category / citation_authorItem _citation.country / _citation.journal_abbrev ... _citation.country / _citation.journal_abbrev / _citation.journal_id_CSD / _citation.journal_id_ISSN / _citation.journal_volume / _citation.page_first / _citation.page_last / _citation.pdbx_database_id_DOI / _citation.pdbx_database_id_PubMed / _citation.title / _citation.year / _citation_author.identifier_ORCID / _citation_author.name Revision 1.2 May 1, 2024 Group / Refinement descriptionCategory / chem_comp_bond / pdbx_initial_refinement_modelRevision 1.3 Nov 20, 2024 Group / Category / Item
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT /
MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 1.45 Å
molecular replacement / Resolution: 1.45 Å  Authors
Authors Switzerland, 3items
Switzerland, 3items  Citation
Citation Journal: Nat Commun / Year: 2021
Journal: Nat Commun / Year: 2021 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 7b4u.cif.gz
7b4u.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb7b4u.ent.gz
pdb7b4u.ent.gz PDB format
PDB format 7b4u.json.gz
7b4u.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/b4/7b4u
https://data.pdbj.org/pub/pdb/validation_reports/b4/7b4u ftp://data.pdbj.org/pub/pdb/validation_reports/b4/7b4u
ftp://data.pdbj.org/pub/pdb/validation_reports/b4/7b4u





 Links
Links Assembly
Assembly


 Components
Components
 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  SLS
SLS  / Beamline: X06SA / Wavelength: 1.00004 Å
/ Beamline: X06SA / Wavelength: 1.00004 Å molecular replacement
molecular replacement Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller











 PDBj
PDBj





