[English] 日本語
 Yorodumi
Yorodumi- PDB-7b2b: Solution structure of a non-covalent extended docking domain comp... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7b2b | ||||||
|---|---|---|---|---|---|---|---|
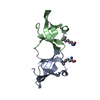
| Title | Solution structure of a non-covalent extended docking domain complex of the Pax NRPS: PaxA T1-CDD/PaxB NDD | ||||||
 Components Components |
| ||||||
 Keywords Keywords | PROTEIN BINDING / PROTEIN | ||||||
| Function / homology |  Function and homology information Function and homology informationphenylalanine racemase (ATP-hydrolysing) / phenylalanine racemase (ATP-hydrolyzing) activity / amino acid activation for nonribosomal peptide biosynthetic process / secondary metabolite biosynthetic process / catalytic activity / phosphopantetheine binding / cytoplasm / cytosol Similarity search - Function | ||||||
| Biological species |  Xenorhabdus cabanillasii (bacteria) Xenorhabdus cabanillasii (bacteria) Xenorhabdus cabanillasii JM26 (bacteria) Xenorhabdus cabanillasii JM26 (bacteria) | ||||||
| Method | SOLUTION NMR / torsion angle dynamics / molecular dynamics | ||||||
 Authors Authors | Watzel, J. / Sarawi, S. / Duchardt-Ferner, E. / Bode, H.B. / Woehnert, J. | ||||||
 Citation Citation |  Journal: Angew.Chem.Int.Ed.Engl. / Year: 2021 Journal: Angew.Chem.Int.Ed.Engl. / Year: 2021Title: Cooperation between a T Domain and a Minimal C-Terminal Docking Domain to Enable Specific Assembly in a Multiprotein NRPS. Authors: Watzel, J. / Duchardt-Ferner, E. / Sarawi, S. / Bode, H.B. / Wohnert, J. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7b2b.cif.gz 7b2b.cif.gz | 965.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7b2b.ent.gz pdb7b2b.ent.gz | 821.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7b2b.json.gz 7b2b.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  7b2b_validation.pdf.gz 7b2b_validation.pdf.gz | 563.7 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  7b2b_full_validation.pdf.gz 7b2b_full_validation.pdf.gz | 922.6 KB | Display | |
| Data in XML |  7b2b_validation.xml.gz 7b2b_validation.xml.gz | 61.1 KB | Display | |
| Data in CIF |  7b2b_validation.cif.gz 7b2b_validation.cif.gz | 91.4 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/b2/7b2b https://data.pdbj.org/pub/pdb/validation_reports/b2/7b2b ftp://data.pdbj.org/pub/pdb/validation_reports/b2/7b2b ftp://data.pdbj.org/pub/pdb/validation_reports/b2/7b2b | HTTPS FTP |
-Related structure data
| Related structure data |  7b2fC C: citing same article ( |
|---|---|
| Similar structure data | |
| Other databases |
|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide | Mass: 3616.222 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Details: a non-native tyrosine residue was added to the C-terminus of the PaxB NDD for an accurate determination of the protein sample concentration Source: (gene. exp.)  Xenorhabdus cabanillasii (bacteria) / Gene: BDD26_3339 / Plasmid: pET-11a / Production host: Xenorhabdus cabanillasii (bacteria) / Gene: BDD26_3339 / Plasmid: pET-11a / Production host:  |
|---|---|
| #2: Protein | Mass: 11976.369 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Details: non-covalent complex of PaxA T1-Cdd 981-1084/PaxB Ndd 1-30 Source: (gene. exp.)  Xenorhabdus cabanillasii JM26 (bacteria) Xenorhabdus cabanillasii JM26 (bacteria)Gene: paxA, XCR1_180002 / Plasmid: pET-11a / Production host:  References: UniProt: W1J0W9, phenylalanine racemase (ATP-hydrolysing) |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
|
 Movie
Movie Controller
Controller











 PDBj
PDBj



 gel filtration
gel filtration