[English] 日本語
 Yorodumi
Yorodumi- PDB-6yxf: Cryogenic human adiponectin receptor 2 (ADIPOR2) with Gd-DO3 liga... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6yxf | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryogenic human adiponectin receptor 2 (ADIPOR2) with Gd-DO3 ligand determined by Serial Crystallography (SSX) using CrystalDirect | ||||||||||||
 Components Components |
| ||||||||||||
 Keywords Keywords | MEMBRANE PROTEIN / Adiponectin receptor / ADIPOR2 / serial synchrotron crystallography / SSX / ligand soaking / CrystalDirect / LCP crystallization / in meso / membrane proteins | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationadiponectin binding / adiponectin-activated signaling pathway / adipokinetic hormone receptor activity / AMPK inhibits chREBP transcriptional activation activity / vascular wound healing / fatty acid oxidation / hormone-mediated signaling pathway / signaling receptor activity / glucose homeostasis / positive regulation of cold-induced thermogenesis ...adiponectin binding / adiponectin-activated signaling pathway / adipokinetic hormone receptor activity / AMPK inhibits chREBP transcriptional activation activity / vascular wound healing / fatty acid oxidation / hormone-mediated signaling pathway / signaling receptor activity / glucose homeostasis / positive regulation of cold-induced thermogenesis / metal ion binding / identical protein binding / plasma membrane Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 3.025 Å MOLECULAR REPLACEMENT / Resolution: 3.025 Å | ||||||||||||
 Authors Authors | Healey, R.D. / Basu, S. / Humm, A.S. / Leyrat, C. / Dupeux, F. / Pica, A. / Granier, S. / Marquez, J.A. | ||||||||||||
| Funding support | European Union, 3items
| ||||||||||||
 Citation Citation |  Journal: Cell Rep Methods / Year: 2021 Journal: Cell Rep Methods / Year: 2021Title: An automated platform for structural analysis of membrane proteins through serial crystallography. Authors: Healey, R.D. / Basu, S. / Humm, A.S. / Leyrat, C. / Cong, X. / Golebiowski, J. / Dupeux, F. / Pica, A. / Granier, S. / Marquez, J.A. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6yxf.cif.gz 6yxf.cif.gz | 129.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6yxf.ent.gz pdb6yxf.ent.gz | 97 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6yxf.json.gz 6yxf.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/yx/6yxf https://data.pdbj.org/pub/pdb/validation_reports/yx/6yxf ftp://data.pdbj.org/pub/pdb/validation_reports/yx/6yxf ftp://data.pdbj.org/pub/pdb/validation_reports/yx/6yxf | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6yx9C  6yxdC  6yxgC  6yxhC  5lwyS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein / Antibody , 2 types, 2 molecules AH
| #1: Protein | Mass: 33097.793 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: ADIPOR2, PAQR2 / Production host: Homo sapiens (human) / Gene: ADIPOR2, PAQR2 / Production host:  |
|---|---|
| #2: Antibody | Mass: 24904.604 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Production host: Homo sapiens (human) / Production host:  |
-Non-polymers , 6 types, 58 molecules 

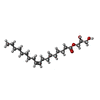

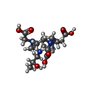






| #3: Chemical | ChemComp-ZN / | ||||||
|---|---|---|---|---|---|---|---|
| #4: Chemical | ChemComp-OLA / | ||||||
| #5: Chemical | ChemComp-OLB / ( #6: Chemical | #7: Chemical | #8: Water | ChemComp-HOH / | |
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.6 Å3/Da / Density % sol: 65.84 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: lipidic cubic phase Details: 30 or 50 nl bolus overlaid with 600 nl precipitant solution using either a Mosquito LCP robot (SPT Labtech) in CrystalDirect plate-2. The crystallisation condition is made of 42.3% PEG 400, ...Details: 30 or 50 nl bolus overlaid with 600 nl precipitant solution using either a Mosquito LCP robot (SPT Labtech) in CrystalDirect plate-2. The crystallisation condition is made of 42.3% PEG 400, 110 mM potassium citrate, 100 mM HEPES pH 7.0. 60-120 nL of Gd-DO3 soaking solution with a final conc. of 10 - 25 mM was delivered onto LCP crystallisation drop. Crystallisation experiments were carried out at the HTX facility of EMBL Grenoble. LCP bolus were harvested automatically at cryogenic condition using CrystalDirect technology. |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: Y |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SLS SLS  / Beamline: X06SA / Wavelength: 1.72 Å / Beamline: X06SA / Wavelength: 1.72 Å |
| Detector | Type: DECTRIS EIGER X 16M / Detector: PIXEL / Date: Nov 22, 2019 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.72 Å / Relative weight: 1 |
| Reflection | Resolution: 3→50 Å / Num. obs: 31468 / % possible obs: 100 % / Redundancy: 14.98 % / CC1/2: 0.98 / Net I/σ(I): 5.06 |
| Reflection shell | Resolution: 3.01→3.1 Å / Num. unique obs: 2336 / CC1/2: 0.3 |
| Serial crystallography sample delivery | Method: fixed target |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 5LWY Resolution: 3.025→48.67 Å / Cor.coef. Fo:Fc: 0.826 / Cor.coef. Fo:Fc free: 0.82 / Cross valid method: THROUGHOUT / σ(F): 0 / SU Rfree Blow DPI: 0.394
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 300 Å2 / Biso mean: 81.24 Å2 / Biso min: 36.47 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.46 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 3.025→48.67 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 3.025→3.06 Å / Rfactor Rfree error: 0
|
 Movie
Movie Controller
Controller












 PDBj
PDBj










