+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6y90 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
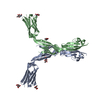
| Title | Structure of full-length CD20 in complex with Rituximab Fab | |||||||||||||||
 Components Components |
| |||||||||||||||
 Keywords Keywords | MEMBRANE PROTEIN / cancer immunotherapy / therapeutic antibody | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationstore-operated calcium entry / calcium ion import into cytosol / positive regulation of calcium ion import across plasma membrane / epidermal growth factor receptor binding / B cell activation / humoral immune response / B cell proliferation / immunoglobulin binding / plasma membrane raft / B cell differentiation ...store-operated calcium entry / calcium ion import into cytosol / positive regulation of calcium ion import across plasma membrane / epidermal growth factor receptor binding / B cell activation / humoral immune response / B cell proliferation / immunoglobulin binding / plasma membrane raft / B cell differentiation / B cell receptor signaling pathway / response to bacterium / protein tetramerization / MHC class II protein complex binding / cell surface receptor signaling pathway / external side of plasma membrane / cell surface / extracellular space / extracellular exosome / nucleoplasm / identical protein binding / plasma membrane Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.69 Å | |||||||||||||||
 Authors Authors | Kumar, A. / Reyes, N. | |||||||||||||||
| Funding support |  France, 1items France, 1items
| |||||||||||||||
 Citation Citation |  Journal: Science / Year: 2020 Journal: Science / Year: 2020Title: Binding mechanisms of therapeutic antibodies to human CD20. Authors: Anand Kumar / Cyril Planchais / Rémi Fronzes / Hugo Mouquet / Nicolas Reyes /  Abstract: Monoclonal antibodies (mAbs) targeting human antigen CD20 (cluster of differentiation 20) constitute important immunotherapies for the treatment of B cell malignancies and autoimmune diseases. Type I ...Monoclonal antibodies (mAbs) targeting human antigen CD20 (cluster of differentiation 20) constitute important immunotherapies for the treatment of B cell malignancies and autoimmune diseases. Type I and II therapeutic mAbs differ in B cell binding properties and cytotoxic effects, reflecting differential interaction mechanisms with CD20. Here we present 3.7- to 4.7-angstrom cryo-electron microscopy structures of full-length CD20 in complexes with prototypical type I rituximab and ofatumumab and type II obinutuzumab. The structures and binding thermodynamics demonstrate that upon binding to CD20, type II mAbs form terminal complexes that preclude recruitment of additional mAbs and complement components, whereas type I complexes act as molecular seeds to increase mAb local concentration for efficient complement activation. Among type I mAbs, ofatumumab complexes display optimal geometry for complement recruitment. The uncovered mechanisms should aid rational design of next-generation immunotherapies targeting CD20. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6y90.cif.gz 6y90.cif.gz | 228.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6y90.ent.gz pdb6y90.ent.gz | 182.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6y90.json.gz 6y90.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/y9/6y90 https://data.pdbj.org/pub/pdb/validation_reports/y9/6y90 ftp://data.pdbj.org/pub/pdb/validation_reports/y9/6y90 ftp://data.pdbj.org/pub/pdb/validation_reports/y9/6y90 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  10731MC  6y92C  6y97C  6y9aC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Protein , 1 types, 2 molecules AB
| #1: Protein | Mass: 19164.797 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Details: Full-length wild type Human CD20 / Source: (gene. exp.)  Homo sapiens (human) / Cell: B-Lymphocyte / Gene: MS4A1, CD20 / Plasmid: pcDNA3.1+ / Cell line (production host): HEK-293F / Production host: Homo sapiens (human) / Cell: B-Lymphocyte / Gene: MS4A1, CD20 / Plasmid: pcDNA3.1+ / Cell line (production host): HEK-293F / Production host:  Homo sapiens (human) / References: UniProt: P11836 Homo sapiens (human) / References: UniProt: P11836 |
|---|
-Antibody , 2 types, 4 molecules CHDL
| #2: Antibody | Mass: 23733.541 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) Homo sapiens (human)#3: Antibody | Mass: 23078.623 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Details: Rituximab Light Chain / Source: (gene. exp.)   Homo sapiens (human) Homo sapiens (human) |
|---|
-Non-polymers , 3 types, 20 molecules 




| #4: Chemical | ChemComp-Y01 / #5: Chemical | #6: Chemical | ChemComp-MYS / |
|---|
-Details
| Has ligand of interest | N |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component |
| |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight |
| |||||||||||||||||||||||||
| Source (natural) |
| |||||||||||||||||||||||||
| Source (recombinant) |
| |||||||||||||||||||||||||
| Buffer solution | pH: 7.4 | |||||||||||||||||||||||||
| Buffer component |
| |||||||||||||||||||||||||
| Specimen | Conc.: 1.5 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES | |||||||||||||||||||||||||
| Specimen support | Grid material: GOLD / Grid mesh size: 300 divisions/in. / Grid type: Quantifoil R1.2/1.3 | |||||||||||||||||||||||||
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 277.15 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Calibrated defocus min: 800 nm / Calibrated defocus max: 2000 nm / Cs: 2.7 mm / C2 aperture diameter: 50 µm |
| Specimen holder | Cryogen: NITROGEN |
| Image recording | Average exposure time: 8 sec. / Electron dose: 41.3 e/Å2 / Detector mode: COUNTING / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Num. of grids imaged: 1 / Num. of real images: 9263 |
| Image scans | Movie frames/image: 40 / Used frames/image: 1-40 |
- Processing
Processing
| Software | Name: PHENIX / Version: 1.18.2_3874: / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 3103137 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Point symmetry: C2 (2 fold cyclic) | ||||||||||||||||||||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3.69 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 65203 / Num. of class averages: 1 / Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Atomic model building | Protocol: AB INITIO MODEL / Space: REAL Details: The CD20-Rituximab fab model was built by placing CD20 peptide (residues 167-186) and Rituximab fab from PDB 2OSL into the EM map. The initial model was fitted manually and extended to a ...Details: The CD20-Rituximab fab model was built by placing CD20 peptide (residues 167-186) and Rituximab fab from PDB 2OSL into the EM map. The initial model was fitted manually and extended to a full CD20 model encompassing residues 45-216. | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Atomic model building | 3D fitting-ID: 1 / Accession code: 2OSL / Initial refinement model-ID: 1 / PDB-ID: 2OSL / Source name: PDB / Type: experimental model
| ||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | Cross valid method: NONE Stereochemistry target values: GeoStd + Monomer Library + CDL v1.2 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 72.58 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj







