[English] 日本語
 Yorodumi
Yorodumi- PDB-6xrz: The 28-kDa Frameshift Stimulation Element from the SARS-CoV-2 RNA... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6xrz | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | The 28-kDa Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome | ||||||||||||||||||||||||
 Components Components | Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome | ||||||||||||||||||||||||
 Keywords Keywords | RNA / Frameshift Stimulation Element / SARS-CoV-2 / COVID-19 | ||||||||||||||||||||||||
| Function / homology | : / RNA / RNA (> 10) Function and homology information Function and homology information | ||||||||||||||||||||||||
| Biological species |  | ||||||||||||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 6.9 Å | ||||||||||||||||||||||||
 Authors Authors | Zhang, K. / Zheludev, I. / Hagey, R. / Wu, M. / Haslecker, R. / Hou, Y. / Kretsch, R. / Pintilie, G. / Rangan, R. / Kladwang, W. ...Zhang, K. / Zheludev, I. / Hagey, R. / Wu, M. / Haslecker, R. / Hou, Y. / Kretsch, R. / Pintilie, G. / Rangan, R. / Kladwang, W. / Li, S. / Pham, E. / Souibgui, C. / Baric, R. / Sheahan, T. / Souza, V. / Glenn, J. / Chiu, W. / Das, R. | ||||||||||||||||||||||||
| Funding support |  United States, 7items United States, 7items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: bioRxiv / Year: 2020 Journal: bioRxiv / Year: 2020Title: Cryo-electron Microscopy and Exploratory Antisense Targeting of the 28-kDa Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome. Authors: Kaiming Zhang / Ivan N Zheludev / Rachel J Hagey / Marie Teng-Pei Wu / Raphael Haslecker / Yixuan J Hou / Rachael Kretsch / Grigore D Pintilie / Ramya Rangan / Wipapat Kladwang / Shanshan Li ...Authors: Kaiming Zhang / Ivan N Zheludev / Rachel J Hagey / Marie Teng-Pei Wu / Raphael Haslecker / Yixuan J Hou / Rachael Kretsch / Grigore D Pintilie / Ramya Rangan / Wipapat Kladwang / Shanshan Li / Edward A Pham / Claire Bernardin-Souibgui / Ralph S Baric / Timothy P Sheahan / Victoria D Souza / Jeffrey S Glenn / Wah Chiu / Rhiju Das Abstract: Drug discovery campaigns against Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) are beginning to target the viral RNA genome . The frameshift stimulation element (FSE) of the SARS-CoV- ...Drug discovery campaigns against Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) are beginning to target the viral RNA genome . The frameshift stimulation element (FSE) of the SARS-CoV-2 genome is required for balanced expression of essential viral proteins and is highly conserved, making it a potential candidate for antiviral targeting by small molecules and oligonucleotides . To aid global efforts focusing on SARS-CoV-2 frameshifting, we report exploratory results from frameshifting and cellular replication experiments with locked nucleic acid (LNA) antisense oligonucleotides (ASOs), which support the FSE as a therapeutic target but highlight difficulties in achieving strong inactivation. To understand current limitations, we applied cryogenic electron microscopy (cryo-EM) and the Ribosolve pipeline to determine a three-dimensional structure of the SARS-CoV-2 FSE, validated through an RNA nanostructure tagging method. This is the smallest macromolecule (88 nt; 28 kDa) resolved by single-particle cryo-EM at subnanometer resolution to date. The tertiary structure model, defined to an estimated accuracy of 5.9 Å, presents a topologically complex fold in which the 5' end threads through a ring formed inside a three-stem pseudoknot. Our results suggest an updated model for SARS-CoV-2 frameshifting as well as binding sites that may be targeted by next generation ASOs and small molecules. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6xrz.cif.gz 6xrz.cif.gz | 531.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6xrz.ent.gz pdb6xrz.ent.gz | 459.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6xrz.json.gz 6xrz.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/xr/6xrz https://data.pdbj.org/pub/pdb/validation_reports/xr/6xrz ftp://data.pdbj.org/pub/pdb/validation_reports/xr/6xrz ftp://data.pdbj.org/pub/pdb/validation_reports/xr/6xrz | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  22296MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 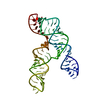
|
|---|---|
| 1 |
|
| Number of models | 10 |
- Components
Components
| #1: RNA chain | Mass: 28285.660 Da / Num. of mol.: 1 / Fragment: GB residues 13434-13521 / Source method: obtained synthetically Source: (synth.)  References: GenBank: 1860584649 |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: 28-kDa Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome Type: COMPLEX / Entity ID: all / Source: NATURAL |
|---|---|
| Molecular weight | Value: 0.028 MDa / Experimental value: NO |
| Source (natural) | Organism:  |
| Buffer solution | pH: 8 |
| Specimen | Conc.: 0.15 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Specimen support | Grid material: COPPER / Grid mesh size: 200 divisions/in. / Grid type: Quantifoil R2/1 |
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Cs: 2.7 mm / C2 aperture diameter: 70 µm |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Average exposure time: 6 sec. / Electron dose: 8.3 e/Å2 / Detector mode: COUNTING / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Num. of real images: 10222 |
| Image scans | Movie frames/image: 30 |
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||
| Symmetry | Point symmetry: C1 (asymmetric) | ||||||||||||||||||||||||
| 3D reconstruction | Resolution: 6.9 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 109137 / Symmetry type: POINT |
 Movie
Movie Controller
Controller










 PDBj
PDBj





























