[English] 日本語
 Yorodumi
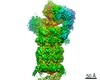
Yorodumi- PDB-6wjd: SA-like state of human 26S Proteasome with non-cleavable M1-linke... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6wjd | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | SA-like state of human 26S Proteasome with non-cleavable M1-linked hexaubiquitin and E3 ubiquitin ligase E6AP/UBE3A | ||||||||||||
 Components Components |
| ||||||||||||
 Keywords Keywords | HYDROLASE/PROTEIN BINDING / 26S protease / ubiquitin / complex / subunit / HYDROLASE-PROTEIN BINDING complex | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationthyrotropin-releasing hormone receptor binding / Impaired BRCA2 translocation to the nucleus / Impaired BRCA2 binding to SEM1 (DSS1) / nuclear proteasome complex / host-mediated perturbation of viral transcription / positive regulation of inclusion body assembly / Hydrolases; Acting on peptide bonds (peptidases); Omega peptidases / proteasome accessory complex / integrator complex / meiosis I ...thyrotropin-releasing hormone receptor binding / Impaired BRCA2 translocation to the nucleus / Impaired BRCA2 binding to SEM1 (DSS1) / nuclear proteasome complex / host-mediated perturbation of viral transcription / positive regulation of inclusion body assembly / Hydrolases; Acting on peptide bonds (peptidases); Omega peptidases / proteasome accessory complex / integrator complex / meiosis I / purine ribonucleoside triphosphate binding / Antigen processing: Ub, ATP-independent proteasomal degradation / proteasome regulatory particle / cytosolic proteasome complex / positive regulation of proteasomal protein catabolic process / proteasome-activating activity / proteasome regulatory particle, lid subcomplex / proteasome regulatory particle, base subcomplex / protein K63-linked deubiquitination / negative regulation of programmed cell death / Regulation of ornithine decarboxylase (ODC) / metal-dependent deubiquitinase activity / Proteasome assembly / Cross-presentation of soluble exogenous antigens (endosomes) / proteasome core complex / Homologous DNA Pairing and Strand Exchange / Defective homologous recombination repair (HRR) due to BRCA1 loss of function / Defective HDR through Homologous Recombination Repair (HRR) due to PALB2 loss of BRCA1 binding function / Defective HDR through Homologous Recombination Repair (HRR) due to PALB2 loss of BRCA2/RAD51/RAD51C binding function / Resolution of D-loop Structures through Synthesis-Dependent Strand Annealing (SDSA) / Somitogenesis / K63-linked deubiquitinase activity / Resolution of D-loop Structures through Holliday Junction Intermediates / transcription factor binding / proteasome binding / Impaired BRCA2 binding to RAD51 / regulation of protein catabolic process / myofibril / proteasome storage granule / Presynaptic phase of homologous DNA pairing and strand exchange / general transcription initiation factor binding / polyubiquitin modification-dependent protein binding / protein deubiquitination / blastocyst development / immune system process / proteasome endopeptidase complex / NF-kappaB binding / proteasome core complex, beta-subunit complex / endopeptidase activator activity / threonine-type endopeptidase activity / proteasome assembly / proteasome core complex, alpha-subunit complex / mRNA export from nucleus / enzyme regulator activity / regulation of proteasomal protein catabolic process / inclusion body / Maturation of protein E / Maturation of protein E / ER Quality Control Compartment (ERQC) / Myoclonic epilepsy of Lafora / FLT3 signaling by CBL mutants / Constitutive Signaling by NOTCH1 HD Domain Mutants / IRAK2 mediated activation of TAK1 complex / Prevention of phagosomal-lysosomal fusion / Alpha-protein kinase 1 signaling pathway / Glycogen synthesis / IRAK1 recruits IKK complex / IRAK1 recruits IKK complex upon TLR7/8 or 9 stimulation / Endosomal Sorting Complex Required For Transport (ESCRT) / Membrane binding and targetting of GAG proteins / Negative regulation of FLT3 / Regulation of TBK1, IKKε (IKBKE)-mediated activation of IRF3, IRF7 / PTK6 Regulates RTKs and Their Effectors AKT1 and DOK1 / Regulation of TBK1, IKKε-mediated activation of IRF3, IRF7 upon TLR3 ligation / IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation / NOTCH2 Activation and Transmission of Signal to the Nucleus / TICAM1,TRAF6-dependent induction of TAK1 complex / TICAM1-dependent activation of IRF3/IRF7 / APC/C:Cdc20 mediated degradation of Cyclin B / proteasome complex / Regulation of FZD by ubiquitination / Downregulation of ERBB4 signaling / TBP-class protein binding / APC-Cdc20 mediated degradation of Nek2A / : / p75NTR recruits signalling complexes / InlA-mediated entry of Listeria monocytogenes into host cells / TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling / Regulation of pyruvate metabolism / NF-kB is activated and signals survival / TRAF6-mediated induction of TAK1 complex within TLR4 complex / Downregulation of ERBB2:ERBB3 signaling / Regulation of innate immune responses to cytosolic DNA / Pexophagy / NRIF signals cell death from the nucleus / Activated NOTCH1 Transmits Signal to the Nucleus / Regulation of PTEN localization / VLDLR internalisation and degradation / Synthesis of active ubiquitin: roles of E1 and E2 enzymes / TICAM1, RIP1-mediated IKK complex recruitment Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 4.8 Å | ||||||||||||
 Authors Authors | Chen, X. / Walters, K.J. | ||||||||||||
| Funding support |  United States, 3items United States, 3items
| ||||||||||||
 Citation Citation |  Journal: Structure / Year: 2020 Journal: Structure / Year: 2020Title: Cryo-EM Reveals Unanchored M1-Ubiquitin Chain Binding at hRpn11 of the 26S Proteasome. Authors: Xiang Chen / Zachary Dorris / Dan Shi / Rick K Huang / Htet Khant / Tara Fox / Natalia de Val / Dewight Williams / Ping Zhang / Kylie J Walters /  Abstract: The 26S proteasome is specialized for regulated protein degradation and formed by a dynamic regulatory particle (RP) that caps a hollow cylindrical core particle (CP) where substrates are proteolyzed. ...The 26S proteasome is specialized for regulated protein degradation and formed by a dynamic regulatory particle (RP) that caps a hollow cylindrical core particle (CP) where substrates are proteolyzed. Its diverse substrates unify as proteasome targets by ubiquitination. We used cryogenic electron microscopy (cryo-EM) to study how human 26S proteasome interacts with M1-linked hexaubiquitin (M1-Ub) unanchored to a substrate and E3 ubiquitin ligase E6AP/UBE3A. Proteasome structures are available with model substrates extending through the RP ATPase ring and substrate-conjugated K63-linked ubiquitin chains present at inhibited deubiquitinating enzyme hRpn11 and the nearby ATPase hRpt4/hRpt5 coiled coil. In this study, we find M1-Ub at the hRpn11 site despite the absence of conjugated substrate, indicating that ubiquitin binding at this location does not require substrate interaction with the RP. Moreover, unanchored M1-Ub binds to this hRpn11 site of the proteasome with the CP gating residues in both the closed and opened conformational states. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6wjd.cif.gz 6wjd.cif.gz | 2.2 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6wjd.ent.gz pdb6wjd.ent.gz | Display |  PDB format PDB format | |
| PDBx/mmJSON format |  6wjd.json.gz 6wjd.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/wj/6wjd https://data.pdbj.org/pub/pdb/validation_reports/wj/6wjd ftp://data.pdbj.org/pub/pdb/validation_reports/wj/6wjd ftp://data.pdbj.org/pub/pdb/validation_reports/wj/6wjd | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  21691MC  6wjnC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10401 (Title: SA-like and SD-like states of human 26S Proteasome with non-cleavable M1-linked hexaubiquitin and E3 ubiquitin ligase E6AP/UBE3A EMPIAR-10401 (Title: SA-like and SD-like states of human 26S Proteasome with non-cleavable M1-linked hexaubiquitin and E3 ubiquitin ligase E6AP/UBE3AData size: 330.3 Data #1: Motion-corrected single frame micrographs of human 26S Proteasome with non-cleavable M1-linked hexaubiquitin and E3 ubiquitin ligase E6AP/UBE3A [micrographs - single frame]) |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-26S proteasome non-ATPase regulatory subunit ... , 11 types, 11 molecules UVWXYZabcdf
| #1: Protein | Mass: 105958.234 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) / References: UniProt: Q99460 Homo sapiens (human) / References: UniProt: Q99460 |
|---|---|
| #2: Protein | Mass: 61066.500 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) / References: UniProt: O43242 Homo sapiens (human) / References: UniProt: O43242 |
| #3: Protein | Mass: 52979.359 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) / References: UniProt: O00232 Homo sapiens (human) / References: UniProt: O00232 |
| #4: Protein | Mass: 47526.688 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) / References: UniProt: O00231 Homo sapiens (human) / References: UniProt: O00231 |
| #5: Protein | Mass: 45592.285 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) / References: UniProt: Q15008 Homo sapiens (human) / References: UniProt: Q15008 |
| #6: Protein | Mass: 37086.441 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) / References: UniProt: P51665 Homo sapiens (human) / References: UniProt: P51665 |
| #7: Protein | Mass: 42995.359 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) / References: UniProt: Q9UNM6 Homo sapiens (human) / References: UniProt: Q9UNM6 |
| #8: Protein | Mass: 40781.590 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) / References: UniProt: P55036 Homo sapiens (human) / References: UniProt: P55036 |
| #9: Protein | Mass: 34488.824 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) Homo sapiens (human)References: UniProt: O00487, Hydrolases; Acting on peptide bonds (peptidases); Omega peptidases |
| #10: Protein | Mass: 39536.676 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) / References: UniProt: P48556 Homo sapiens (human) / References: UniProt: P48556 |
| #12: Protein | Mass: 100313.625 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) / References: UniProt: Q13200 Homo sapiens (human) / References: UniProt: Q13200 |
-Protein , 2 types, 2 molecules eu
| #11: Protein | Mass: 8284.611 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) / References: UniProt: P60896 Homo sapiens (human) / References: UniProt: P60896 |
|---|---|
| #19: Protein | Mass: 8576.831 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) / References: UniProt: P0CG48 Homo sapiens (human) / References: UniProt: P0CG48 |
-26S proteasome regulatory subunit ... , 6 types, 6 molecules ABCDEF
| #13: Protein | Mass: 48700.805 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) / References: UniProt: P35998 Homo sapiens (human) / References: UniProt: P35998 |
|---|---|
| #14: Protein | Mass: 49260.504 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) / References: UniProt: P62191 Homo sapiens (human) / References: UniProt: P62191 |
| #15: Protein | Mass: 44852.121 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) / References: UniProt: P62195 Homo sapiens (human) / References: UniProt: P62195 |
| #16: Protein | Mass: 47426.141 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) / References: UniProt: P43686 Homo sapiens (human) / References: UniProt: P43686 |
| #17: Protein | Mass: 45867.027 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) / References: UniProt: A0A087X2I1 Homo sapiens (human) / References: UniProt: A0A087X2I1 |
| #18: Protein | Mass: 49266.457 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) / References: UniProt: P17980 Homo sapiens (human) / References: UniProt: P17980 |
-Proteasome subunit alpha type- ... , 7 types, 14 molecules GgHhIiJjKkLlMm
| #20: Protein | Mass: 27301.262 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) Homo sapiens (human)References: UniProt: P60900, proteasome endopeptidase complex #21: Protein | Mass: 25796.338 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) Homo sapiens (human)References: UniProt: P25787, proteasome endopeptidase complex #22: Protein | Mass: 29394.648 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) Homo sapiens (human)References: UniProt: P25789, proteasome endopeptidase complex #23: Protein | Mass: 27798.695 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) Homo sapiens (human)References: UniProt: O14818, proteasome endopeptidase complex #24: Protein | Mass: 26304.779 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) Homo sapiens (human)References: UniProt: P28066, proteasome endopeptidase complex #25: Protein | Mass: 30150.277 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) Homo sapiens (human)References: UniProt: P25786, proteasome endopeptidase complex #26: Protein | Mass: 28338.057 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) Homo sapiens (human)References: UniProt: P25788, proteasome endopeptidase complex |
|---|
-Proteasome subunit beta type- ... , 7 types, 14 molecules NnOoPpQqRrSsTt
| #27: Protein | Mass: 25246.455 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) Homo sapiens (human)References: UniProt: P28072, proteasome endopeptidase complex #28: Protein | Mass: 29869.223 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) Homo sapiens (human)References: UniProt: Q99436, proteasome endopeptidase complex #29: Protein | Mass: 22841.701 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) Homo sapiens (human)References: UniProt: P49720, proteasome endopeptidase complex #30: Protein | Mass: 22864.277 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) Homo sapiens (human)References: UniProt: P49721, proteasome endopeptidase complex #31: Protein | Mass: 28379.053 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) Homo sapiens (human)References: UniProt: P28074, proteasome endopeptidase complex #32: Protein | Mass: 26391.201 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) Homo sapiens (human)References: UniProt: P20618, proteasome endopeptidase complex #33: Protein | Mass: 29099.986 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) Homo sapiens (human)References: UniProt: P28070, proteasome endopeptidase complex |
|---|
-Non-polymers , 4 types, 12 molecules 






| #34: Chemical | ChemComp-ZN / | ||||
|---|---|---|---|---|---|
| #35: Chemical | ChemComp-AGS / #36: Chemical | ChemComp-MG / #37: Chemical | |
-Details
| Has ligand of interest | N |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Human 26S Proteasome with non-cleavable M1-linked hexaubiquitin and E3 ubiquitin ligase E6AP/UBE3A Type: COMPLEX / Entity ID: #1-#33 / Source: NATURAL | |||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||||||||||||||||
| Buffer solution | pH: 7.5 Details: 50 mM Tris, pH 7.5, 50 mM NaCl, 1.5 mM ATP-gamma-S, 5 mM MgCl2, 2 mM DTT, 10 uM zinc sulfate | |||||||||||||||||||||||||||||||||||
| Buffer component |
| |||||||||||||||||||||||||||||||||||
| Specimen | Conc.: 1 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES | |||||||||||||||||||||||||||||||||||
| Specimen support | Grid material: COPPER / Grid mesh size: 200 divisions/in. / Grid type: Quantifoil R1.2/1.3 | |||||||||||||||||||||||||||||||||||
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 291.15 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Average exposure time: 0.2 sec. / Electron dose: 36.64 e/Å2 / Detector mode: SUPER-RESOLUTION / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Num. of grids imaged: 1 / Num. of real images: 6216 |
| Image scans | Movie frames/image: 40 / Used frames/image: 1-40 |
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 1050369 | ||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Point symmetry: C1 (asymmetric) | ||||||||||||||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 4.8 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 37698 / Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||||||||||
| Atomic model building | Protocol: RIGID BODY FIT / Space: REAL | ||||||||||||||||||||||||||||||||||||||||||||
| Atomic model building | PDB-ID: 6MSD Accession code: 6MSD / Source name: PDB / Type: experimental model |
 Movie
Movie Controller
Controller

















 PDBj
PDBj



























