| Entry | Database: PDB / ID: 6wa3
|
|---|
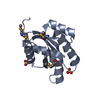
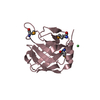
| Title | Solution NMR structure of the myristoylated feline immunodeficiency virus matrix protein |
|---|
 Components Components | Matrix protein |
|---|
 Keywords Keywords | VIRAL PROTEIN / FIV |
|---|
| Function / homology |  Function and homology information Function and homology information
viral budding via host ESCRT complex / viral capsid / viral nucleocapsid / structural constituent of virion / nucleic acid binding / zinc ion bindingSimilarity search - Function : / Matrix protein, lentiviral and alpha-retroviral, N-terminal / Retroviral nucleocapsid Gag protein p24, C-terminal domain / Gag protein p24 C-terminal domain / Retrovirus capsid, C-terminal / Retrovirus capsid, N-terminal / zinc finger / Zinc knuckle / Zinc finger, CCHC-type superfamily / Zinc finger, CCHC-type / Zinc finger CCHC-type profile.Similarity search - Domain/homology |
|---|
| Biological species |  Feline immunodeficiency virus Feline immunodeficiency virus |
|---|
| Method | SOLUTION NMR / na |
|---|
 Authors Authors | Brown, J.B. / Summers, H.R. / Brown, L.A. / Marchant, J. / Canova, P.N. / O'Hern, C.T. / Abbott, S. / Nyaunu, C. / Maxwell, S. / Johnson, T. ...Brown, J.B. / Summers, H.R. / Brown, L.A. / Marchant, J. / Canova, P.N. / O'Hern, C.T. / Abbott, S. / Nyaunu, C. / Maxwell, S. / Johnson, T. / Moser, M. / Ablan, S.D. / Carter, H. / Freed, E.O. / Summers, M.F. |
|---|
| Funding support |  United States, 9items United States, 9items | Organization | Grant number | Country |
|---|
| Howard Hughes Medical Institute (HHMI) | |  United States United States | | National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | T32 GM066706 |  United States United States | | National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | R01 GM042561 |  United States United States | | National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | IMSD R25 GM-055036-18 |  United States United States | | National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | U54 GM103297 |  United States United States | | National Science Foundation (NSF, United States) | DGE-1144243 |  United States United States | | National Science Foundation (NSF, United States) | 1460653 |  United States United States | | National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | R37 AI30917 |  United States United States | | National Institutes of Health/National Cancer Institute (NIH/NCI) | |  United States United States |
|
|---|
 Citation Citation |  Journal: J.Mol.Biol. / Year: 2020 Journal: J.Mol.Biol. / Year: 2020
Title: Structural and Mechanistic Studies of the Rare Myristoylation Signal of the Feline Immunodeficiency Virus.
Authors: Brown, J.B. / Summers, H.R. / Brown, L.A. / Marchant, J. / Canova, P.N. / O'Hern, C.T. / Abbott, S.T. / Nyaunu, C. / Maxwell, S. / Johnson, T. / Moser, M.B. / Ablan, S.D. / Carter, H. / ...Authors: Brown, J.B. / Summers, H.R. / Brown, L.A. / Marchant, J. / Canova, P.N. / O'Hern, C.T. / Abbott, S.T. / Nyaunu, C. / Maxwell, S. / Johnson, T. / Moser, M.B. / Ablan, S.D. / Carter, H. / Freed, E.O. / Summers, M.F. |
|---|
| History | | Deposition | Mar 24, 2020 | Deposition site: RCSB / Processing site: RCSB |
|---|
| Revision 1.0 | Jul 22, 2020 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Dec 21, 2022 | Group: Database references / Category: database_2 / struct_ref_seq_dif
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _struct_ref_seq_dif.details |
|---|
| Revision 2.0 | May 17, 2023 | Group: Advisory / Atomic model ...Advisory / Atomic model / Database references / Derived calculations / Polymer sequence / Source and taxonomy / Structure summary
Category: atom_site / entity ...atom_site / entity / entity_name_com / entity_poly / entity_poly_seq / entity_src_gen / pdbx_poly_seq_scheme / pdbx_unobs_or_zero_occ_residues / struct_conf / struct_conn / struct_ref_seq / struct_ref_seq_dif / struct_site_gen
Item: _atom_site.auth_atom_id / _atom_site.label_atom_id ..._atom_site.auth_atom_id / _atom_site.label_atom_id / _atom_site.label_seq_id / _entity.formula_weight / _entity_poly.nstd_monomer / _entity_poly.pdbx_seq_one_letter_code / _entity_poly.pdbx_seq_one_letter_code_can / _entity_src_gen.pdbx_end_seq_num / _struct_conf.beg_label_seq_id / _struct_conf.end_label_seq_id / _struct_conn.ptnr1_label_seq_id / _struct_ref_seq.seq_align_beg / _struct_ref_seq.seq_align_end / _struct_site_gen.label_seq_id |
|---|
| Revision 2.1 | Jun 14, 2023 | Group: Other / Category: pdbx_database_status / Item: _pdbx_database_status.status_code_nmr_data |
|---|
| Revision 2.2 | Nov 6, 2024 | Group: Data collection / Database references / Structure summary
Category: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_entry_details / pdbx_modification_feature
Item: _database_2.pdbx_DOI / _pdbx_entry_details.has_protein_modification |
|---|
|
|---|
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Feline immunodeficiency virus
Feline immunodeficiency virus Authors
Authors United States, 9items
United States, 9items  Citation
Citation Journal: J.Mol.Biol. / Year: 2020
Journal: J.Mol.Biol. / Year: 2020 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 6wa3.cif.gz
6wa3.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb6wa3.ent.gz
pdb6wa3.ent.gz PDB format
PDB format 6wa3.json.gz
6wa3.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/wa/6wa3
https://data.pdbj.org/pub/pdb/validation_reports/wa/6wa3 ftp://data.pdbj.org/pub/pdb/validation_reports/wa/6wa3
ftp://data.pdbj.org/pub/pdb/validation_reports/wa/6wa3

 Links
Links Assembly
Assembly
 Components
Components Feline immunodeficiency virus / Gene: GAG / Production host:
Feline immunodeficiency virus / Gene: GAG / Production host: 
 Sample preparation
Sample preparation Movie
Movie Controller
Controller










 PDBj
PDBj






 HSQC
HSQC