+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6r6d | |||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
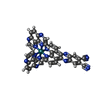
| Title | [Ru(TAP)2(11,12-CN2-dppz)]2+ bound to d(TCGGCGCCGA)2 | |||||||||||||||||||||||||||||||
 Components Components | DNA (5'-D(* Keywords KeywordsDNA / Ruthenium / intercalation / asymmetric | Function / homology | : / Chem-EQQ / DNA |  Function and homology information Function and homology informationBiological species | synthetic construct (others) | Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  SAD / Resolution: 1.838 Å SAD / Resolution: 1.838 Å  Authors AuthorsMcQuaid, K.T. / Hall, J.P. / Cardin, C.J. | Funding support | |  United Kingdom, 2items United Kingdom, 2items
 Citation Citation Journal: Chemistry / Year: 2018 Journal: Chemistry / Year: 2018Title: X-ray Crystal Structures Show DNA Stacking Advantage of Terminal Nitrile Substitution in Ru-dppz Complexes. Authors: McQuaid, K. / Hall, J.P. / Brazier, J.A. / Cardin, D.J. / Cardin, C.J. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6r6d.cif.gz 6r6d.cif.gz | 28 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6r6d.ent.gz pdb6r6d.ent.gz | 17.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6r6d.json.gz 6r6d.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/r6/6r6d https://data.pdbj.org/pub/pdb/validation_reports/r6/6r6d ftp://data.pdbj.org/pub/pdb/validation_reports/r6/6r6d ftp://data.pdbj.org/pub/pdb/validation_reports/r6/6r6d | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 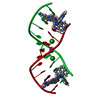
| ||||||||
| Unit cell |
|
- Components
Components
| #1: DNA chain | Mass: 3045.992 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.) synthetic construct (others) |
|---|---|
| #2: Chemical | ChemComp-BA / |
| #3: Chemical | ChemComp-EQQ / |
| #4: Chemical | ChemComp-NA / |
| #5: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.07 Å3/Da / Density % sol: 59.95 % / Description: Orthorhombic |
|---|---|
| Crystal grow | Temperature: 292 K / Method: vapor diffusion, sitting drop / pH: 7 Details: 1uL 1mM dsDNA, 1ul 5mM rac[Ru(TAP)2(11,12-CN2-dppz)]2+, 6ul of a solution containing 10% (v/v) 2-methyl-2,4-pentanediol, 80mM NaCl, 20mM Na-cacodylate pH 7, 20mM BaCl2 and 12mM spermine ...Details: 1uL 1mM dsDNA, 1ul 5mM rac[Ru(TAP)2(11,12-CN2-dppz)]2+, 6ul of a solution containing 10% (v/v) 2-methyl-2,4-pentanediol, 80mM NaCl, 20mM Na-cacodylate pH 7, 20mM BaCl2 and 12mM spermine tetra-HCl equilibrated against 500uL 35% 2-methyl-2,4-pentanediol. |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Diamond Diamond  / Beamline: I03 / Wavelength: 0.9763 Å / Beamline: I03 / Wavelength: 0.9763 Å |
| Detector | Type: DECTRIS PILATUS3 S 6M / Detector: PIXEL / Date: Feb 18, 2019 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9763 Å / Relative weight: 1 |
| Reflection | Resolution: 1.838→34.31 Å / Num. obs: 6316 / % possible obs: 100 % / Redundancy: 23.5 % / CC1/2: 0.999 / Rmerge(I) obs: 0.097 / Rpim(I) all: 0.021 / Rrim(I) all: 0.1 / Net I/σ(I): 17.7 |
| Reflection shell | Resolution: 1.84→1.87 Å / Redundancy: 24.5 % / Num. unique obs: 180 / % possible all: 98.9 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  SAD / Resolution: 1.838→33.025 Å / SU ML: 0.2 / Cross valid method: FREE R-VALUE / σ(F): 1.34 / Phase error: 26.54 SAD / Resolution: 1.838→33.025 Å / SU ML: 0.2 / Cross valid method: FREE R-VALUE / σ(F): 1.34 / Phase error: 26.54
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.838→33.025 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller











 PDBj
PDBj