| Entry | Database: PDB / ID: 6ohh
|
|---|
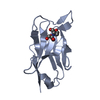
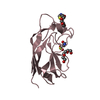
| Title | Structure of EF1p2_mFAP2b bound to DFHBI |
|---|
 Components Components | EF1p2_mFAP2b |
|---|
 Keywords Keywords | DE NOVO PROTEIN / Rossetta / ligand binder / computational / beta barrel / EF-motif |
|---|
| Function / homology | Chem-38E Function and homology information Function and homology information |
|---|
| Biological species | synthetic construct (others) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 2.1 Å molecular replacement / Resolution: 2.1 Å |
|---|
 Authors Authors | Doyle, L.A. / Stoddard, B.L. |
|---|
| Funding support |  United States, 1items United States, 1items | Organization | Grant number | Country |
|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | R01 GM115545 |  United States United States |
|
|---|
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021
Title: Incorporation of sensing modalities into de novo designed fluorescence-activating proteins
Authors: Klima, J.C. / Doyle, L.A. / Lee, J.D. / Rappleye, M. / Gagnon, L.A. / Lee, M.Y. / Barros, E.P. / Vorobieva, A.A. / Dou, J. / Bremner, S. / Jacob, S. / Quon, J.S. / Chow, C.M. / Carter, L. / ...Authors: Klima, J.C. / Doyle, L.A. / Lee, J.D. / Rappleye, M. / Gagnon, L.A. / Lee, M.Y. / Barros, E.P. / Vorobieva, A.A. / Dou, J. / Bremner, S. / Jacob, S. / Quon, J.S. / Chow, C.M. / Carter, L. / Mack, D.L. / Amaro, R.E. / Vaughan, J.C. / Berndt, A. / Stoddard, B.L. / Baker, D. |
|---|
| History | | Deposition | Apr 5, 2019 | Deposition site: RCSB / Processing site: RCSB |
|---|
| Revision 1.0 | Apr 8, 2020 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Feb 17, 2021 | Group: Database references / Derived calculations / Category: citation / citation_author / struct_conn
Item: _citation.country / _citation.journal_abbrev ..._citation.country / _citation.journal_abbrev / _citation.journal_id_CSD / _citation.journal_id_ISSN / _citation.journal_volume / _citation.page_first / _citation.pdbx_database_id_DOI / _citation.title / _citation.year / _struct_conn.pdbx_dist_value / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id |
|---|
| Revision 1.2 | Mar 13, 2024 | Group: Data collection / Database references / Refinement description
Category: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / struct_ncs_dom_lim
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession ..._database_2.pdbx_DOI / _database_2.pdbx_database_accession / _struct_ncs_dom_lim.beg_auth_comp_id / _struct_ncs_dom_lim.beg_label_asym_id / _struct_ncs_dom_lim.beg_label_comp_id / _struct_ncs_dom_lim.beg_label_seq_id / _struct_ncs_dom_lim.end_auth_comp_id / _struct_ncs_dom_lim.end_label_asym_id / _struct_ncs_dom_lim.end_label_comp_id / _struct_ncs_dom_lim.end_label_seq_id |
|---|
|
|---|
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT /
MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 2.1 Å
molecular replacement / Resolution: 2.1 Å  Authors
Authors United States, 1items
United States, 1items  Citation
Citation Journal: Nat Commun / Year: 2021
Journal: Nat Commun / Year: 2021 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 6ohh.cif.gz
6ohh.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb6ohh.ent.gz
pdb6ohh.ent.gz PDB format
PDB format 6ohh.json.gz
6ohh.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/oh/6ohh
https://data.pdbj.org/pub/pdb/validation_reports/oh/6ohh ftp://data.pdbj.org/pub/pdb/validation_reports/oh/6ohh
ftp://data.pdbj.org/pub/pdb/validation_reports/oh/6ohh Links
Links Assembly
Assembly


 Movie
Movie Controller
Controller











 PDBj
PDBj
