Entry Database : PDB / ID : 6lu7Title The crystal structure of COVID-19 main protease in complex with an inhibitor N3 3C-like proteinase N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE Keywords / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species synthetic construct (others) Method / / / Resolution : 2.16 Å Authors Liu, X. / Zhang, B. / Jin, Z. / Yang, H. / Rao, Z. Journal : Nature / Year : 2020Title : Structure of Mprofrom SARS-CoV-2 and discovery of its inhibitors.
Authors: Jin, Z. / Du, X. / Xu, Y. / Deng, Y. / Liu, M. / Zhao, Y. / Zhang, B. / Li, X. / Zhang, L. / Peng, C. / Duan, Y. / Yu, J. / Wang, L. / Yang, K. / Liu, F. / Jiang, R. / Yang, X. / You, T. / ... Authors : Jin, Z. / Du, X. / Xu, Y. / Deng, Y. / Liu, M. / Zhao, Y. / Zhang, B. / Li, X. / Zhang, L. / Peng, C. / Duan, Y. / Yu, J. / Wang, L. / Yang, K. / Liu, F. / Jiang, R. / Yang, X. / You, T. / Liu, X. / Yang, X. / Bai, F. / Liu, H. / Liu, X. / Guddat, L.W. / Xu, W. / Xiao, G. / Qin, C. / Shi, Z. / Jiang, H. / Rao, Z. / Yang, H. #1: Journal : Biorxiv / Year : 2020Title : Structure of Mpro from COVID-19 virus and discovery of its inhibitors.
Authors: Jin, Z. / Du, X. / Xu, Y. / Deng, Y. / Liu, M. / Zhao, Y. / Zhang, B. / Li, X. / Zhang, L. / Peng, C. / Duan, Y. / Yu, J. / Wang, L. / Yang, K. / Liu, F. / Jiang, R. / Yang, X. / You, T. / ... Authors : Jin, Z. / Du, X. / Xu, Y. / Deng, Y. / Liu, M. / Zhao, Y. / Zhang, B. / Li, X. / Zhang, L. / Peng, C. / Duan, Y. / Yu, J. / Wang, L. / Yang, K. / Liu, F. / Jiang, R. / Yang, X. / You, T. / Liu, X. / Yang, X. / Bai, F. / Liu, H. / Liu, X. / Guddat, L. / Xu, W. / Xiao, G. / Qin, C. / Shi, Z. / Jiang, H. / Rao, Z. / Yang, H. History Deposition Jan 26, 2020 Deposition site / Processing site Revision 1.0 Feb 5, 2020 Provider / Type Revision 2.0 Feb 12, 2020 Group Advisory / Atomic model ... Advisory / Atomic model / Data collection / Database references / Derived calculations / Refinement description / Structure summary Category atom_site / citation ... atom_site / citation / entity / pdbx_nonpoly_scheme / pdbx_struct_assembly_prop / pdbx_struct_sheet_hbond / pdbx_struct_special_symmetry / pdbx_validate_rmsd_bond / pdbx_validate_symm_contact / pdbx_validate_torsion / refine / refine_hist / refine_ls_shell / software / struct / struct_conn / struct_site / struct_site_gen Item _citation.title / _entity.pdbx_number_of_molecules ... _citation.title / _entity.pdbx_number_of_molecules / _pdbx_struct_assembly_prop.value / _pdbx_struct_sheet_hbond.range_1_auth_comp_id / _pdbx_struct_sheet_hbond.range_1_auth_seq_id / _pdbx_struct_sheet_hbond.range_1_label_comp_id / _pdbx_struct_sheet_hbond.range_1_label_seq_id / _pdbx_struct_sheet_hbond.range_2_auth_comp_id / _pdbx_struct_sheet_hbond.range_2_auth_seq_id / _pdbx_struct_sheet_hbond.range_2_label_comp_id / _pdbx_struct_sheet_hbond.range_2_label_seq_id / _pdbx_validate_rmsd_bond.bond_deviation / _pdbx_validate_rmsd_bond.bond_value / _pdbx_validate_torsion.phi / _pdbx_validate_torsion.psi / _refine.B_iso_max / _refine.B_iso_mean / _refine.B_iso_min / _refine.ls_R_factor_R_free / _refine.ls_R_factor_R_work / _refine.ls_R_factor_obs / _refine.ls_number_reflns_R_work / _refine.overall_SU_ML / _refine.pdbx_overall_phase_error / _refine.pdbx_stereochemistry_target_values / _refine.solvent_model_details / _refine_hist.number_atoms_solvent / _refine_hist.number_atoms_total / _refine_hist.pdbx_B_iso_mean_ligand / _refine_hist.pdbx_B_iso_mean_solvent / _refine_ls_shell.R_factor_R_free / _refine_ls_shell.R_factor_R_work / _software.classification / _software.name / _software.version / _struct.title / _struct_conn.pdbx_dist_value Description / Provider / Type Revision 2.1 Feb 19, 2020 Group / Structure summary / Category / structItem / _citation.title / _struct.titleRevision 2.2 Feb 26, 2020 Group / Category / Item Revision 2.3 Mar 11, 2020 Group / Structure summary / Category / entity_src_gen / structItem _entity.pdbx_description / _entity_src_gen.gene_src_common_name ... _entity.pdbx_description / _entity_src_gen.gene_src_common_name / _entity_src_gen.pdbx_gene_src_ncbi_taxonomy_id / _entity_src_gen.pdbx_gene_src_scientific_name / _struct.pdbx_descriptor Revision 2.4 Mar 18, 2020 Group / Category / citation_authorItem _citation.country / _citation.journal_abbrev ... _citation.country / _citation.journal_abbrev / _citation.journal_id_CSD / _citation.pdbx_database_id_DOI / _citation.title / _citation.year Revision 2.5 Apr 22, 2020 Group / Category / citation_authorRevision 2.6 May 6, 2020 Group / Source and taxonomy / Structure summaryCategory entity / entity_name_com ... entity / entity_name_com / entity_src_gen / struct / struct_ref / struct_ref_seq Item _entity.pdbx_description / _entity.pdbx_ec ... _entity.pdbx_description / _entity.pdbx_ec / _entity_src_gen.gene_src_common_name / _entity_src_gen.pdbx_gene_src_gene / _struct.pdbx_descriptor / _struct_ref.db_code / _struct_ref.db_name / _struct_ref.pdbx_align_begin / _struct_ref.pdbx_db_accession / _struct_ref.pdbx_seq_one_letter_code / _struct_ref_seq.db_align_beg / _struct_ref_seq.db_align_end / _struct_ref_seq.pdbx_db_accession Revision 2.7 May 27, 2020 Group / Category / citation_author / Item / _citation_author.identifier_ORCIDRevision 2.8 Jun 10, 2020 Group / Category / Item Revision 2.9 Jun 24, 2020 Group / Category Item / _citation.page_first / _citation.page_lastRevision 3.0 Jul 29, 2020 Group Advisory / Atomic model ... Advisory / Atomic model / Author supporting evidence / Data collection / Database references / Derived calculations / Refinement description / Structure summary Category atom_site / citation_author ... atom_site / citation_author / entity / pdbx_entity_instance_feature / pdbx_nonpoly_scheme / pdbx_poly_seq_scheme / pdbx_struct_assembly_prop / pdbx_validate_rmsd_bond / pdbx_validate_symm_contact / refine / refine_hist / reflns_shell / struct_conn Item _atom_site.B_iso_or_equiv / _atom_site.Cartn_x ... _atom_site.B_iso_or_equiv / _atom_site.Cartn_x / _atom_site.Cartn_y / _atom_site.Cartn_z / _atom_site.occupancy / _citation_author.identifier_ORCID / _entity.pdbx_fragment / _pdbx_nonpoly_scheme.auth_seq_num / _pdbx_poly_seq_scheme.auth_mon_id / _pdbx_poly_seq_scheme.auth_seq_num / _pdbx_struct_assembly_prop.value / _refine.B_iso_mean / _refine.ls_R_factor_obs / _refine.ls_percent_reflns_obs / _refine.pdbx_starting_model / _refine_hist.pdbx_B_iso_mean_ligand / _refine_hist.pdbx_B_iso_mean_solvent / _refine_hist.pdbx_number_atoms_ligand / _refine_hist.pdbx_number_atoms_protein / _refine_hist.pdbx_number_residues_total / _struct_conn.pdbx_dist_value Description / Provider / Type Revision 3.1 Mar 10, 2021 Group / Category / entity_name_com / Item / _entity_name_com.nameRevision 4.0 Nov 15, 2023 Group Atomic model / Data collection ... Atomic model / Data collection / Database references / Derived calculations Category atom_site / chem_comp_atom ... atom_site / chem_comp_atom / chem_comp_bond / citation / database_2 / pdbx_validate_rmsd_angle / struct_conn Item _atom_site.auth_atom_id / _atom_site.label_atom_id ... _atom_site.auth_atom_id / _atom_site.label_atom_id / _citation.journal_id_ISSN / _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr2_label_atom_id Revision 4.1 Nov 29, 2023 Group / Category Revision 4.2 Nov 20, 2024 Group / Category / pdbx_modification_feature / Item
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.16 Å
MOLECULAR REPLACEMENT / Resolution: 2.16 Å  Authors
Authors Citation
Citation Journal: Nature / Year: 2020
Journal: Nature / Year: 2020 Journal: Biorxiv / Year: 2020
Journal: Biorxiv / Year: 2020 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 6lu7.cif.gz
6lu7.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb6lu7.ent.gz
pdb6lu7.ent.gz PDB format
PDB format 6lu7.json.gz
6lu7.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/lu/6lu7
https://data.pdbj.org/pub/pdb/validation_reports/lu/6lu7 ftp://data.pdbj.org/pub/pdb/validation_reports/lu/6lu7
ftp://data.pdbj.org/pub/pdb/validation_reports/lu/6lu7

 Links
Links Assembly
Assembly

 Components
Components

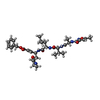
 Type: Peptide-like / Class: Inhibitor / Mass: 680.791 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.) synthetic construct (others)
Type: Peptide-like / Class: Inhibitor / Mass: 680.791 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.) synthetic construct (others) X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  SSRF
SSRF  / Beamline: BL17U1 / Wavelength: 1.0718 Å
/ Beamline: BL17U1 / Wavelength: 1.0718 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller






















 PDBj
PDBj









