| 登録情報 | データベース: PDB / ID: 6k3a
|
|---|
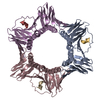
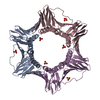
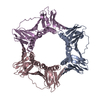
| タイトル | Crystal structure of human PCNA in complex with DNMT1 PIP box motif. |
|---|
 要素 要素 | - Peptide from DNA (cytosine-5)-methyltransferase 1
- Proliferating cell nuclear antigen
|
|---|
 キーワード キーワード | REPLICATION / DNMT1 / PCNA / DNA methylation / PIP box |
|---|
| 機能・相同性 |  機能・相同性情報 機能・相同性情報
chromosomal DNA methylation maintenance following DNA replication / negative regulation of vascular associated smooth muscle cell differentiation involved in phenotypic switching / epigenetic programming of gene expression / cellular response to bisphenol A / DNA-methyltransferase activity / positive regulation of deoxyribonuclease activity / dinucleotide insertion or deletion binding / negative regulation of vascular associated smooth muscle cell apoptotic process / PCNA-p21 complex / mitotic telomere maintenance via semi-conservative replication ...chromosomal DNA methylation maintenance following DNA replication / negative regulation of vascular associated smooth muscle cell differentiation involved in phenotypic switching / epigenetic programming of gene expression / cellular response to bisphenol A / DNA-methyltransferase activity / positive regulation of deoxyribonuclease activity / dinucleotide insertion or deletion binding / negative regulation of vascular associated smooth muscle cell apoptotic process / PCNA-p21 complex / mitotic telomere maintenance via semi-conservative replication / purine-specific mismatch base pair DNA N-glycosylase activity / MutLalpha complex binding / positive regulation of DNA-directed DNA polymerase activity / nuclear lamina / DNA (cytosine-5-)-methyltransferase / DNA (cytosine-5-)-methyltransferase activity / Polymerase switching / Telomere C-strand (Lagging Strand) Synthesis / Processive synthesis on the lagging strand / PCNA complex / SUMOylation of DNA methylation proteins / Removal of the Flap Intermediate / Processive synthesis on the C-strand of the telomere / Mismatch repair (MMR) directed by MSH2:MSH3 (MutSbeta) / Polymerase switching on the C-strand of the telomere / Mismatch repair (MMR) directed by MSH2:MSH6 (MutSalpha) / replisome / STAT3 nuclear events downstream of ALK signaling / Transcription of E2F targets under negative control by DREAM complex / Removal of the Flap Intermediate from the C-strand / female germ cell nucleus / methyl-CpG binding / DNA methylation-dependent constitutive heterochromatin formation / response to L-glutamate / negative regulation of gene expression via chromosomal CpG island methylation / histone acetyltransferase binding / leading strand elongation / DNA polymerase processivity factor activity / G1/S-Specific Transcription / response to dexamethasone / replication fork processing / nuclear replication fork / SUMOylation of DNA replication proteins / PCNA-Dependent Long Patch Base Excision Repair / estrous cycle / mismatch repair / translesion synthesis / pericentric heterochromatin / response to cadmium ion / Nuclear events stimulated by ALK signaling in cancer / DNA polymerase binding / cyclin-dependent protein kinase holoenzyme complex / positive regulation of vascular associated smooth muscle cell proliferation / epithelial cell differentiation / base-excision repair, gap-filling / positive regulation of DNA repair / Translesion synthesis by REV1 / Translesion synthesis by POLK / DNA methylation / TP53 Regulates Transcription of Genes Involved in G2 Cell Cycle Arrest / Translesion synthesis by POLI / Gap-filling DNA repair synthesis and ligation in GG-NER / positive regulation of DNA replication / PRC2 methylates histones and DNA / male germ cell nucleus / replication fork / Defective pyroptosis / nuclear estrogen receptor binding / promoter-specific chromatin binding / liver regeneration / cellular response to amino acid stimulus / Recognition of DNA damage by PCNA-containing replication complex / Termination of translesion DNA synthesis / Translesion Synthesis by POLH / NoRC negatively regulates rRNA expression / HDR through Homologous Recombination (HRR) / Dual Incision in GG-NER / receptor tyrosine kinase binding / cellular response to hydrogen peroxide / Dual incision in TC-NER / Gap-filling DNA repair synthesis and ligation in TC-NER / cellular response to UV / cellular response to xenobiotic stimulus / E3 ubiquitin ligases ubiquitinate target proteins / response to estradiol / heart development / methylation / damaged DNA binding / chromosome, telomeric region / nuclear body / negative regulation of gene expression / centrosome / DNA-templated transcription / chromatin binding / protein-containing complex binding / positive regulation of gene expression / chromatin / negative regulation of transcription by RNA polymerase II / enzyme binding / DNA binding類似検索 - 分子機能 DNA (cytosine-5)-methyltransferase 1-like / DNA (cytosine-5)-methyltransferase 1, replication foci domain / Cytosine specific DNA methyltransferase replication foci domain / DMAP1-binding Domain / DMAP1-binding Domain / DMAP1-binding domain / DMAP1-binding domain profile. / : / DNA methylase, C-5 cytosine-specific, conserved site / C-5 cytosine-specific DNA methylases C-terminal signature. ...DNA (cytosine-5)-methyltransferase 1-like / DNA (cytosine-5)-methyltransferase 1, replication foci domain / Cytosine specific DNA methyltransferase replication foci domain / DMAP1-binding Domain / DMAP1-binding Domain / DMAP1-binding domain / DMAP1-binding domain profile. / : / DNA methylase, C-5 cytosine-specific, conserved site / C-5 cytosine-specific DNA methylases C-terminal signature. / Box / Proliferating Cell Nuclear Antigen / Proliferating Cell Nuclear Antigen - #10 / CXXC zinc finger domain / Zinc finger, CXXC-type / Zinc finger CXXC-type profile. / DNA methylase, C-5 cytosine-specific, active site / C-5 cytosine-specific DNA methylases active site. / C-5 cytosine-specific DNA methylase (Dnmt) domain profile. / C-5 cytosine methyltransferase / C-5 cytosine-specific DNA methylase / Proliferating cell nuclear antigen signature 2. / Bromo adjacent homology domain / BAH domain / Bromo adjacent homology (BAH) domain / Bromo adjacent homology (BAH) domain superfamily / BAH domain profile. / Proliferating cell nuclear antigen, PCNA, conserved site / Proliferating cell nuclear antigen signature 1. / Proliferating cell nuclear antigen, PCNA / Proliferating cell nuclear antigen, PCNA, N-terminal / Proliferating cell nuclear antigen, PCNA, C-terminal / Proliferating cell nuclear antigen, N-terminal domain / Proliferating cell nuclear antigen, C-terminal domain / : / S-adenosyl-L-methionine-dependent methyltransferase superfamily / Alpha Beta類似検索 - ドメイン・相同性 DNA methyltransferase 1 / Proliferating cell nuclear antigen / DNA (cytosine-5)-methyltransferase 1類似検索 - 構成要素 |
|---|
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) |
|---|
| 手法 |  X線回折 / X線回折 /  シンクロトロン / シンクロトロン /  分子置換 / 解像度: 2.3 Å 分子置換 / 解像度: 2.3 Å |
|---|
 データ登録者 データ登録者 | Jimenji, T. / Kori, S. / Arita, K. |
|---|
| 資金援助 |  日本, 2件 日本, 2件 | 組織 | 認可番号 | 国 |
|---|
| Ministry of Education, Culture, Sports, Science and Technology (Japan) | 18H02392 |  日本 日本 | | Japan Science and Technology | 14530337 |  日本 日本 |
|
|---|
 引用 引用 |  ジャーナル: Biochem.Biophys.Res.Commun. / 年: 2019 ジャーナル: Biochem.Biophys.Res.Commun. / 年: 2019
タイトル: Structure of PCNA in complex with DNMT1 PIP box reveals the basis for the molecular mechanism of the interaction.
著者: Jimenji, T. / Matsumura, R. / Kori, S. / Arita, K. |
|---|
| 履歴 | | 登録 | 2019年5月17日 | 登録サイト: PDBJ / 処理サイト: PDBJ |
|---|
| 改定 1.0 | 2019年6月26日 | Provider: repository / タイプ: Initial release |
|---|
| 改定 1.1 | 2019年7月10日 | Group: Data collection / Database references / カテゴリ: citation
Item: _citation.country / _citation.journal_abbrev ..._citation.country / _citation.journal_abbrev / _citation.journal_id_ASTM / _citation.journal_id_CSD / _citation.journal_id_ISSN / _citation.pdbx_database_id_DOI / _citation.pdbx_database_id_PubMed / _citation.title / _citation.year |
|---|
| 改定 1.2 | 2019年7月24日 | Group: Data collection / Database references / カテゴリ: citation
Item: _citation.journal_volume / _citation.page_first / _citation.page_last |
|---|
| 改定 1.3 | 2023年11月22日 | Group: Data collection / Database references / Refinement description
カテゴリ: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession |
|---|
|
|---|
 データを開く
データを開く 基本情報
基本情報 要素
要素 キーワード
キーワード 機能・相同性情報
機能・相同性情報 Homo sapiens (ヒト)
Homo sapiens (ヒト) X線回折 /
X線回折 /  シンクロトロン /
シンクロトロン /  分子置換 / 解像度: 2.3 Å
分子置換 / 解像度: 2.3 Å  データ登録者
データ登録者 日本, 2件
日本, 2件  引用
引用 ジャーナル: Biochem.Biophys.Res.Commun. / 年: 2019
ジャーナル: Biochem.Biophys.Res.Commun. / 年: 2019 構造の表示
構造の表示 Molmil
Molmil Jmol/JSmol
Jmol/JSmol ダウンロードとリンク
ダウンロードとリンク ダウンロード
ダウンロード 6k3a.cif.gz
6k3a.cif.gz PDBx/mmCIF形式
PDBx/mmCIF形式 pdb6k3a.ent.gz
pdb6k3a.ent.gz PDB形式
PDB形式 6k3a.json.gz
6k3a.json.gz PDBx/mmJSON形式
PDBx/mmJSON形式 その他のダウンロード
その他のダウンロード 6k3a_validation.pdf.gz
6k3a_validation.pdf.gz wwPDB検証レポート
wwPDB検証レポート 6k3a_full_validation.pdf.gz
6k3a_full_validation.pdf.gz 6k3a_validation.xml.gz
6k3a_validation.xml.gz 6k3a_validation.cif.gz
6k3a_validation.cif.gz https://data.pdbj.org/pub/pdb/validation_reports/k3/6k3a
https://data.pdbj.org/pub/pdb/validation_reports/k3/6k3a ftp://data.pdbj.org/pub/pdb/validation_reports/k3/6k3a
ftp://data.pdbj.org/pub/pdb/validation_reports/k3/6k3a
 リンク
リンク 集合体
集合体
 要素
要素 Homo sapiens (ヒト) / 遺伝子: PCNA / 発現宿主:
Homo sapiens (ヒト) / 遺伝子: PCNA / 発現宿主: 
 Homo sapiens (ヒト) / 参照: UniProt: K7ERQ1, UniProt: P26358*PLUS
Homo sapiens (ヒト) / 参照: UniProt: K7ERQ1, UniProt: P26358*PLUS X線回折 / 使用した結晶の数: 1
X線回折 / 使用した結晶の数: 1  試料調製
試料調製 シンクロトロン / サイト:
シンクロトロン / サイト:  Photon Factory
Photon Factory  / ビームライン: BL-17A / 波長: 0.98 Å
/ ビームライン: BL-17A / 波長: 0.98 Å 解析
解析 分子置換
分子置換 ムービー
ムービー コントローラー
コントローラー










 PDBj
PDBj


















